Detection of head-to-tail DNA sequences of human bocavirus in clinical samples
- PMID: 21573237
- PMCID: PMC3087758
- DOI: 10.1371/journal.pone.0019457
Detection of head-to-tail DNA sequences of human bocavirus in clinical samples
Abstract
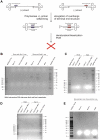
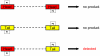
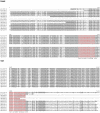
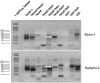
Parvoviruses are single stranded DNA viruses that replicate in a so called "rolling-hairpin" mechanism, a variant of the rolling circle replication known for bacteriophages like φX174. The replication intermediates of parvoviruses thus are concatemers of head-to-head or tail-to-tail structure. Surprisingly, in case of the novel human bocavirus, neither head-to-head nor tail-to-tail DNA sequences were detected in clinical isolates; in contrast head-to-tail DNA sequences were identified by PCR and sequencing. Thereby, the head-to-tail sequences were linked by a novel sequence of 54 bp of which 20 bp also occur as conserved structures of the palindromic ends of parvovirus MVC which in turn is a close relative to human bocavirus.
Conflict of interest statement
Figures
Similar articles
-
Hairpin Transfer-Independent Parvovirus DNA Replication Produces Infectious Virus.J Virol. 2021 Sep 27;95(20):e0110821. doi: 10.1128/JVI.01108-21. Epub 2021 Aug 4. J Virol. 2021. PMID: 34346761 Free PMC article.
-
Diverse Head-to-Tail Sequences in the Circular Genome of Human Bocavirus Genotype 1 among Children with Acute Respiratory Infections Implied the Switch of Template Chain in the Rolling-Circle Replication Model.Pathogens. 2024 Sep 3;13(9):757. doi: 10.3390/pathogens13090757. Pathogens. 2024. PMID: 39338948 Free PMC article.
-
The Human Bocavirus Is Associated with Some Lung and Colorectal Cancers and Persists in Solid Tumors.PLoS One. 2013 Jun 27;8(6):e68020. doi: 10.1371/journal.pone.0068020. Print 2013. PLoS One. 2013. PMID: 23826357 Free PMC article.
-
[Human bocavirus and its current epidemic status in China].Bing Du Xue Bao. 2013 Jan;29(1):56-64. Bing Du Xue Bao. 2013. PMID: 23547381 Review. Chinese.
-
[Progress on development and research of human bocavirus 1].Bing Du Xue Bao. 2014 Jan;30(1):103-8. Bing Du Xue Bao. 2014. PMID: 24772907 Review. Chinese.
Cited by
-
Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia.PLoS Pathog. 2012;8(8):e1002899. doi: 10.1371/journal.ppat.1002899. Epub 2012 Aug 30. PLoS Pathog. 2012. PMID: 22956907 Free PMC article.
-
Characterization of a novel porcine parvovirus tentatively designated PPV5.PLoS One. 2013 Jun 7;8(6):e65312. doi: 10.1371/journal.pone.0065312. Print 2013. PLoS One. 2013. PMID: 23762339 Free PMC article.
-
Fatal HBoV-1 infection in adult female cystic fibrosis patient.Hum Pathol (N Y). 2017 Mar;7:51-52. doi: 10.1016/j.ehpc.2016.07.001. Epub 2016 Jul 18. Hum Pathol (N Y). 2017. PMID: 32337159 Free PMC article.
-
Porcine bocavirus: achievements in the past five years.Viruses. 2014 Dec 10;6(12):4946-60. doi: 10.3390/v6124946. Viruses. 2014. PMID: 25514206 Free PMC article. Review.
-
Detection of spliced mRNA from human bocavirus 1 in clinical samples from children with respiratory tract infections.Emerg Infect Dis. 2013 Apr;19(4):574-80. doi: 10.3201/eid1904.121775. Emerg Infect Dis. 2013. PMID: 23628409 Free PMC article.
References
-
- Tattersall P. Kerr JRB, M.E., Cotmore S, editors. The evolution of Parvovirus Taxonomy. 2006. pp. 5–14.
-
- Tattersall P, Ward DC. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976;263:106–109. - PubMed
-
- Hirt B. Molecular biology of autonomous parvoviruses. Contrib Microbiol. 2000;4:163–177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

