Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity
- PMID: 21575642
- PMCID: PMC3114172
- DOI: 10.1016/j.jmb.2011.04.078
Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity
Abstract
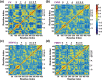
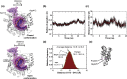
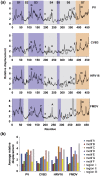
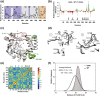
The viral RNA-dependent RNA polymerase (RdRp) is essential for multiplication of all RNA viruses. The sequence diversity of an RNA virus population contributes to its ability to infect the host. This diversity emanates from errors made by the RdRp during RNA synthesis. The physical basis for RdRp fidelity is unclear but is linked to conformational changes occurring during the nucleotide-addition cycle. To understand RdRp dynamics that might influence RdRp function, we have analyzed all-atom molecular dynamics simulations on the nanosecond timescale of four RdRps from the picornavirus family that exhibit 30-74% sequence identity. Principal component analysis showed that the major motions observed during the simulations derived from conserved structural motifs and regions of known function. The dynamics of residues participating in the same biochemical property, for example, RNA binding, nucleotide binding or catalysis, were correlated even when spatially distant on the RdRp structure. The conserved and correlated dynamics of functional structural elements suggest coevolution of dynamics with structure and function of the RdRp. Crystal structures of all picornavirus RdRps exhibit a template-nascent RNA duplex channel too small to fully accommodate duplex RNA. Simulations revealed opening and closing motions of the RNA and nucleoside triphosphate channels, which might be relevant to nucleoside triphosphate entry, inorganic pyrophosphate exit and translocation. A role for nanosecond timescale dynamics in RdRp fidelity is supported by the altered dynamics of the high-fidelity G64S derivative of PV RdRp relative to wild-type enzyme.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures





References
-
- World Health Organization. (2008). Laboratory surveillance for wild and vaccine-derived polioviruses, January 2007–June 2008. Wkly. Epidemiol. Rec.83, 321–328. - PubMed
-
- De Clercq E. The design of drugs for HIV and HCV. Nat. Rev., Drug. Discov. 2007;6:1001–1018. - PubMed
-
- Graci J.D., Cameron C.E. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antiviral Chem. Chemother. 2004;15:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

