ΔNp63 versatilely regulates a Broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation
- PMID: 21576089
- PMCID: PMC3443863
- DOI: 10.1158/0008-5472.CAN-10-3445
ΔNp63 versatilely regulates a Broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation
Erratum in
- Cancer Res. 2011 Dec 1;71(23):7323
Abstract
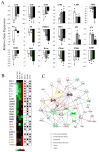
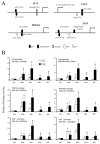
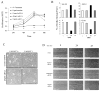
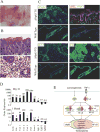
Head and neck squamous cell carcinoma (HNSCC) and many other epithelial malignancies exhibit increased proliferation, invasion, and inflammation, concomitant with aberrant nuclear activation of TP53 and NF-κB family members ΔNp63, cRel, and RelA. However, the mechanisms of cross-talk by which these transcription factors coordinate gene expression and the malignant phenotype remain elusive. In this study, we showed that ΔNp63 regulates a cohort of genes involved in cell growth, survival, adhesion, and inflammation, which substantially overlaps with the NF-κB transcriptome. ΔNp63 with cRel and/or RelA are recruited to form novel binding complexes on p63 or NF-κB/Rel sites of multitarget gene promoters. Overexpressed ΔNp63- or TNF-α-induced NF-κB and inflammatory cytokine interleukin-8 (IL-8) reporter activation depended on RelA/cRel regulatory binding sites. Depletion of RelA or ΔNp63 by small interfering RNA (siRNA) significantly inhibited NF-κB-specific, or TNF-α-induced IL-8 reporter activation. ΔNp63 siRNA significantly inhibited proliferation, survival, and migration by HNSCC cells in vitro. Consistent with these data, an increase in nuclear ΔNp63, accompanied by increased proliferation (Ki-67) and adhesion (β4 integrin) markers, and induced inflammatory cell infiltration was observed throughout HNSCC specimens, when compared with the basilar pattern of protein expression and minimal inflammation seen in nonmalignant mucosa. Furthermore, overexpression of ΔNp63α in squamous epithelial cells in transgenic mice leads to increased suprabasilar cRel, Ki-67, and cytokine expression, together with epidermal hyperplasia and diffuse inflammation, similar to HNSCC. Our study reveals ΔNp63 as a master transcription factor that, in coordination with NF-κB/Rels, orchestrates a broad gene program promoting epidermal hyperplasia, inflammation, and the malignant phenotype of HNSCC.
©2011 AACR
Conflict of interest statement
Figures



References
-
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. - PubMed
-
- Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–8. - PubMed
-
- Dey A, Tergaonkar V, Lane DP. Double-edged swords as cancer therapeutics: simultaneously targeting p53 and NF-kappaB pathways. Nat Rev Drug Discov. 2008;7:1031–40. - PubMed
-
- Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res. 2007;13:1076–82. - PubMed
-
- Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1:493–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

