Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling
- PMID: 21576221
- PMCID: PMC3159477
- DOI: 10.1093/nar/gkr343
Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling
Abstract
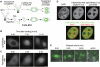
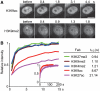
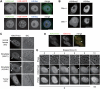
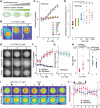
Histone modifications play an important role in epigenetic gene regulation and genome integrity. It remains largely unknown, however, how these modifications dynamically change in individual cells. By using fluorescently labeled specific antigen binding fragments (Fabs), we have developed a general method to monitor the distribution and global level of endogenous histone H3 lysine modifications in living cells without disturbing cell growth and embryo development. Fabs produce distinct nuclear patterns that are characteristic of their target modifications. H3K27 trimethylation-specific Fabs, for example, are concentrated on inactive X chromosomes. As Fabs bind their targets transiently, the ratio of bound and free molecules depends on the target concentration, allowing us to measure changes in global modification levels. High-affinity Fabs are suitable for mouse embryo imaging, so we have used them to monitor H3K9 and H3K27 acetylation levels in mouse preimplantation embryos produced by in vitro fertilization and somatic cell nuclear transfer. The data suggest that a high level of H3K27 acetylation is important for normal embryo development. As Fab-based live endogenous modification labeling (FabLEM) is broadly useful for visualizing any modification, it should be a powerful tool for studying cell signaling and diagnosis in the future.
Figures

References
-
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell. Biol. 2010;11:427–439. - PubMed
-
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. - PubMed
-
- Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006;23:289–296. - PubMed
-
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14 (Spec No 1), R47–R58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

