Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity
- PMID: 21576344
- PMCID: PMC3191967
- DOI: 10.1128/IAI.00097-11
Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity
Abstract
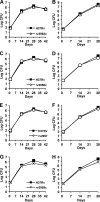
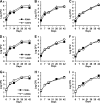
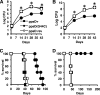
Onset of the adaptive immune response in mice infected with Mycobacterium tuberculosis is accompanied by slowing of bacterial replication and establishment of a chronic infection. Stabilization of bacterial numbers during the chronic phase of infection is dependent on the activity of the gamma interferon (IFN-γ)-inducible nitric oxide synthase (NOS2). Previously, we described a differential signature-tagged mutagenesis screen designed to identify M. tuberculosis "counterimmune" mechanisms and reported the isolation of three mutants in the H37Rv strain background containing transposon insertions in the rv0072, rv0405, and rv2958c genes. These mutants were impaired for replication and virulence in NOS2(-/-) mice but were growth-proficient and virulent in IFN-γ(-/-) mice, suggesting that the disrupted genes were required for bacterial resistance to an IFN-γ-dependent immune mechanism other than NOS2. Here, we report that the attenuation of these strains is attributable to an underlying transposon-independent deficiency in biosynthesis of phthiocerol dimycocerosate (PDIM), a cell wall lipid that is required for full virulence in mice. We performed whole-genome resequencing of a PDIM-deficient clone and identified a spontaneous point mutation in the putative polyketide synthase PpsD that results in a G44C amino acid substitution. We demonstrate by complementation with the wild-type ppsD gene and reversion of the ppsD gene to the wild-type sequence that the ppsD(G44C) point mutation is responsible for PDIM deficiency, virulence attenuation in NOS2(-/-) and wild-type C57BL/6 mice, and a growth advantage in vitro in liquid culture. We conclude that PDIM biosynthesis is required for M. tuberculosis resistance to an IFN-γ-mediated immune response that is independent of NOS2.
Figures



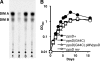
References
-
- Andreu N., Gibert I. 2008. Cell population heterogeneity in Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb.) 88:553–559 - PubMed
-
- Camacho L. R., et al. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845–19854 - PubMed
-
- Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257–267 - PubMed
-
- Constant P., et al. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex: evidence that all strains synthesize glycosylated p-hydroxybenzoic acid methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277:38148–38158 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

