Drosophila histone locus bodies form by hierarchical recruitment of components
- PMID: 21576393
- PMCID: PMC3166876
- DOI: 10.1083/jcb.201012077
Drosophila histone locus bodies form by hierarchical recruitment of components
Abstract
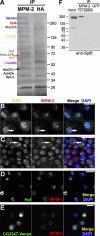
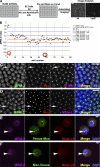
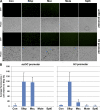
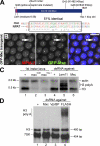
Nuclear bodies are protein- and RNA-containing structures that participate in a wide range of processes critical to genome function. Molecular self-organization is thought to drive nuclear body formation, but whether this occurs stochastically or via an ordered, hierarchical process is not fully understood. We addressed this question using RNAi and proteomic approaches in Drosophila melanogaster to identify and characterize novel components of the histone locus body (HLB), a nuclear body involved in the expression of replication-dependent histone genes. We identified the transcription elongation factor suppressor of Ty 6 (Spt6) and a homologue of mammalian nuclear protein of the ataxia telangiectasia-mutated locus that is encoded by the homeotic gene multisex combs (mxc) as novel HLB components. By combining genetic manipulation in both cell culture and embryos with cytological observations of Mxc, Spt6, and the known HLB components, FLICE-associated huge protein, Mute, U7 small nuclear ribonucleoprotein, and MPM-2 phosphoepitope, we demonstrated sequential recruitment and hierarchical dependency for localization of factors to HLBs during development, suggesting that ordered assembly can play a role in nuclear body formation.
Figures



Comment in
-
Harvey McMahon: ahead of the curve on membrane dynamics.J Cell Biol. 2011 May 16;193(4):598-9. doi: 10.1083/jcb.1934pi. J Cell Biol. 2011. PMID: 21576387 Free PMC article.
Comment on
-
Seed and grow: a two-step model for nuclear body biogenesis.J Cell Biol. 2011 May 16;193(4):605-6. doi: 10.1083/jcb.201104087. J Cell Biol. 2011. PMID: 21576389 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

