Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites
- PMID: 21576453
- PMCID: PMC3147642
- DOI: 10.1128/AAC.01793-10
Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites
Abstract
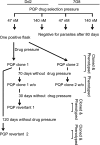
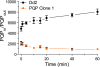
The combination of piperaquine and dihydroartemisinin has recently become the official first-line therapy in several Southeast Asian countries. The pharmacokinetic mismatching of these drugs, whose plasma half-lives are ~20 days and ~1 h, respectively, implies that recrudescent or new infections emerging shortly after treatment cessation will encounter piperaquine as a monotherapy agent. This creates substantial selection pressure for the emergence of resistance. To elucidate potential resistance determinants, we subjected cloned Plasmodium falciparum Dd2 parasites to continuous piperaquine pressure in vitro (47 nM; ~2-fold higher than the Dd2 50% inhibitory concentration [IC(50)]). The phenotype of outgrowth parasites was assayed in two clones, revealing an IC(50) against piperaquine of 2.1 μM and 1.7 μM, over 100-fold greater than that of the parent. To identify the genetic determinant of resistance, we employed comparative whole-genome hybridization analysis. Compared to the Dd2 parent, this analysis found (in both resistant clones) a novel single-nucleotide polymorphism in P. falciparum crt (pfcrt), deamplification of an 82-kb region of chromosome 5 (that includes pfmdr1), and amplification of an adjacent 63-kb region of chromosome 5. Continued propagation without piperaquine selection pressure resulted in "revertant" piperaquine-sensitive parasites. These retained the pfcrt polymorphism and further deamplified the chromosome 5 segment that encompasses pfmdr1; however, these two independently generated revertants both lost the neighboring 63-kb amplification. These results suggest that a copy number variation event on chromosome 5 (825600 to 888300) is associated with piperaquine resistance. Transgene expression studies are underway with individual genes in this segment to evaluate their contribution to piperaquine resistance.
Figures
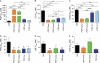

References
-
- Ashley E. A., et al. 2004. Randomized, controlled dose-optimization studies of dihydroartemisinin-piperaquine for the treatment of uncomplicated multidrug-resistant falciparum malaria in Thailand. J. Infect. Dis. 190:1773–1782 - PubMed
-
- Briolant S., Wurtz N., Zettor A., Rogier C., Pradines B. 2010. Susceptibility of Plasmodium falciparum isolates to doxycycline is associated with pftetQ sequence polymorphisms and pftetQ and pfmdt copy numbers. J. Infect. Dis. 201:153–159 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

