Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum
- PMID: 21576455
- PMCID: PMC3107335
- DOI: 10.1073/pnas.1018749108
Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum
Abstract
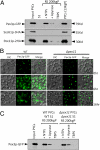
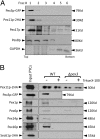
Several yeast and mammalian peroxisomal membrane proteins (PMPs) are delivered to peroxisomes via the endoplasmic reticulum (ER). Fluorescence microscopy showed a focused assembly of PMPs in a specialized domain of the ER, referred to as the preperoxisomal ER. It is proposed that preperoxisomal vesicles containing PMPs bud from this domain to either fuse with preexisting peroxisomes or to mature into functional peroxisomes by uptake of peroxisomal membrane and matrix proteins. However, such vesicular entities are not identified nor are the biochemical requirements for the budding process known. We developed an in vitro cell-free ER-budding assay using Pichia pastoris and followed two endogenous PMPs, Pex11p and Pex3p during their ER exit. Both the PMPs were copackaged in the ER-budded vesicles that float on a Nycodenz gradient. PMP budding from the ER was dependent on ATP, temperature, cytosol, and Pex19p and generated preperoxisomal vesicles with an incomplete complement of PMPs. Surprisingly, Pex11p budding was independent of Pex3p; however, the budded vesicles were devoid of most of the PMPs otherwise present in the wild-type vesicles and might represent peroxisomal remnants. Our findings provide a biochemical platform to uncover the mechanism of PMP budding from the ER.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA. Molecular mechanisms of organelle inheritance: Lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol. 2010;11:644–654. - PubMed
-
- Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: A story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

