Mapping of the minimal inorganic phosphate transporting unit of human PiT2 suggests a structure universal to PiT-related proteins from all kingdoms of life
- PMID: 21586110
- PMCID: PMC3126765
- DOI: 10.1186/1471-2091-12-21
Mapping of the minimal inorganic phosphate transporting unit of human PiT2 suggests a structure universal to PiT-related proteins from all kingdoms of life
Abstract
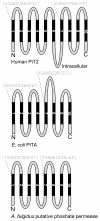
Background: The inorganic (Pi) phosphate transporter (PiT) family comprises known and putative Na(+)- or H(+)-dependent Pi-transporting proteins with representatives from all kingdoms. The mammalian members are placed in the outer cell membranes and suggested to supply cells with Pi to maintain house-keeping functions. Alignment of protein sequences representing PiT family members from all kingdoms reveals the presence of conserved amino acids and that bacterial phosphate permeases and putative phosphate permeases from archaea lack substantial parts of the protein sequence when compared to the mammalian PiT family members. Besides being Na(+)-dependent P(i) (NaP(i)) transporters, the mammalian PiT paralogs, PiT1 and PiT2, also are receptors for gamma-retroviruses. We have here exploited the dual-function of PiT1 and PiT2 to study the structure-function relationship of PiT proteins.
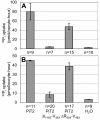
Results: We show that the human PiT2 histidine, H(502), and the human PiT1 glutamate, E(70),--both conserved in eukaryotic PiT family members--are critical for P(i) transport function. Noticeably, human PiT2 H(502) is located in the C-terminal PiT family signature sequence, and human PiT1 E(70) is located in ProDom domains characteristic for all PiT family members.A human PiT2 truncation mutant, which consists of the predicted 10 transmembrane (TM) domain backbone without a large intracellular domain (human PiT2ΔR(254)-V(483)), was found to be a fully functional P(i) transporter. Further truncation of the human PiT2 protein by additional removal of two predicted TM domains together with the large intracellular domain created a mutant that resembles a bacterial phosphate permease and an archaeal putative phosphate permease. This human PiT2 truncation mutant (human PiT2ΔL(183)-V(483)) did also support P(i) transport albeit at very low levels.
Conclusions: The results suggest that the overall structure of the P(i)-transporting unit of the PiT family proteins has remained unchanged during evolution. Moreover, in combination, our studies of the gene structure of the human PiT1 and PiT2 genes (SLC20A1 and SLC20A2, respectively) and alignment of protein sequences of PiT family members from all kingdoms, along with the studies of the dual functions of the human PiT paralogs show that these proteins are excellent as models for studying the evolution of a protein's structure-function relationship.
© 2011 Bøttger and Pedersen; licensee BioMed Central Ltd.
Figures
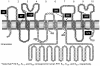



References
-
- Oláh Z, Lehel C, Anderson WB, Eiden MV, Wilson CA. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269(41):25426–25431. - PubMed
-
- Kavanaugh MP, Miller DG, Zhang W, Law W, Kozak SL, Kabat D, Miller AD. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91(15):7071–7075. doi: 10.1073/pnas.91.15.7071. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

