Prediction of peptides binding to the PKA RIIalpha subunit using a hierarchical strategy
- PMID: 21586518
- PMCID: PMC3117377
- DOI: 10.1093/bioinformatics/btr294
Prediction of peptides binding to the PKA RIIalpha subunit using a hierarchical strategy
Abstract
Motivation: Favorable interaction between the regulatory subunit of the cAMP-dependent protein kinase (PKA) and a peptide in A-kinase anchoring proteins (AKAPs) is critical for translocating PKA to the subcellular sites where the enzyme phosphorylates its substrates. It is very hard to identify AKAPs peptides binding to PKA due to the high sequence diversity of AKAPs.
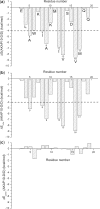
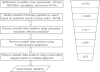
Results: We propose a hierarchical and efficient approach, which combines molecular dynamics (MD) simulations, free energy calculations, virtual mutagenesis (VM) and bioinformatics analyses, to predict peptides binding to the PKA RIIα regulatory subunit in the human proteome systematically. Our approach successfully retrieved 15 out of 18 documented RIIα-binding peptides. Literature curation supported that many newly predicted peptides might be true AKAPs. Here, we present the first systematic search for AKAP peptides in the human proteome, which is useful to further experimental identification of AKAPs and functional analysis of their biological roles.
Figures







Similar articles
-
A systematic evaluation of protein kinase A-A-kinase anchoring protein interaction motifs.Biochemistry. 2015 Jan 13;54(1):11-21. doi: 10.1021/bi500721a. Epub 2014 Sep 10. Biochemistry. 2015. PMID: 25097019
-
AKAP18:PKA-RIIα structure reveals crucial anchor points for recognition of regulatory subunits of PKA.Biochem J. 2016 Jul 1;473(13):1881-94. doi: 10.1042/BCJ20160242. Epub 2016 Apr 21. Biochem J. 2016. PMID: 27102985 Free PMC article.
-
Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding.J Mol Biol. 2000 Apr 28;298(2):329-39. doi: 10.1006/jmbi.2000.3662. J Mol Biol. 2000. PMID: 10764601
-
Compartmentalization role of A-kinase anchoring proteins (AKAPs) in mediating protein kinase A (PKA) signaling and cardiomyocyte hypertrophy.Int J Mol Sci. 2014 Dec 24;16(1):218-29. doi: 10.3390/ijms16010218. Int J Mol Sci. 2014. PMID: 25547489 Free PMC article. Review.
-
Pharmacological Interference With Protein-protein Interactions of Akinase Anchoring Proteins as a Strategy for the Treatment of Disease.Curr Drug Targets. 2016;17(10):1147-71. doi: 10.2174/1389450116666150416114247. Curr Drug Targets. 2016. PMID: 25882214 Review.
Cited by
-
Theoretical Studies on the Selectivity Mechanisms of Glycogen Synthase Kinase 3β (GSK3β) with Pyrazine ATP-competitive Inhibitors by 3DQSAR, Molecular Docking, Molecular Dynamics Simulation and Free Energy Calculations.Curr Comput Aided Drug Des. 2020;16(1):17-30. doi: 10.2174/1573409915666190708102459. Curr Comput Aided Drug Des. 2020. PMID: 31284868 Free PMC article.
-
In Silico Exploration for Novel Type-I Inhibitors of Tie-2/TEK: The Performance of Different Selection Strategy in Selecting Virtual Screening Candidates.Sci Rep. 2016 Nov 23;6:37628. doi: 10.1038/srep37628. Sci Rep. 2016. PMID: 27876862 Free PMC article.
-
LRBA is essential for urinary concentration and body water homeostasis.Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2202125119. doi: 10.1073/pnas.2202125119. Epub 2022 Jul 21. Proc Natl Acad Sci U S A. 2022. PMID: 35862451 Free PMC article.
-
Targeting the actin nucleation promoting factor WASp provides a therapeutic approach for hematopoietic malignancies.Nat Commun. 2021 Sep 22;12(1):5581. doi: 10.1038/s41467-021-25842-7. Nat Commun. 2021. PMID: 34552085 Free PMC article.
-
Lipopolysaccharide-responsive beige-like anchor is involved in regulating NF-κB activation in B cells.Front Immunol. 2024 Jul 15;15:1409434. doi: 10.3389/fimmu.2024.1409434. eCollection 2024. Front Immunol. 2024. PMID: 39076990 Free PMC article.
References
-
- Discovery Studio 2.5 Guide (2009) Accelrys Inc., San Diego.
-
- Alto N.M., Scott J.D. The role of A-kinase anchoring proteins in cAMP-mediated signal transduction pathways. Cell Biochem. Biophys. 2004:201–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

