Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan
- PMID: 21586574
- PMCID: PMC3129176
- DOI: 10.1074/jbc.M111.241414
Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan
Abstract
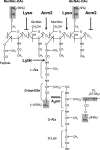
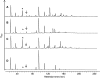
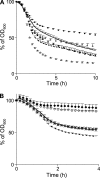
Peptidoglycan (PG) N-acetyl muramic acid (MurNAc) O-acetylation is widely spread in gram-positive bacteria and is generally associated with resistance against lysozyme and endogenous autolysins. We report here the presence of O-acetylation on N-acetylglucosamine (GlcNAc) in Lactobacillus plantarum PG. This modification of glycan strands was never described in bacteria. Fine structural characterization of acetylated muropeptides released from L. plantarum PG demonstrated that both MurNAc and GlcNAc are O-acetylated in this species. These two PG post-modifications rely on two dedicated O-acetyltransferase encoding genes, named oatA and oatB, respectively. By analyzing the resistance to cell wall hydrolysis of mutant strains, we showed that GlcNAc O-acetylation inhibits N-acetylglucosaminidase Acm2, the major L. plantarum autolysin. In this bacterial species, inactivation of oatA, encoding MurNAc O-acetyltransferase, resulted in marked sensitivity to lysozyme. Moreover, MurNAc over-O-acetylation was shown to activate autolysis through the putative N-acetylmuramoyl-L-alanine amidase LytH enzyme. Our data indicate that in L. plantarum, two different O-acetyltransferases play original and antagonistic roles in the modulation of the activity of endogenous autolysins.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

