Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages
- PMID: 21587207
- PMCID: PMC3155298
- DOI: 10.1038/emboj.2011.163
Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages
Abstract
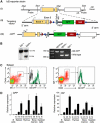
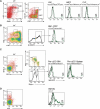
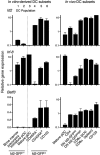
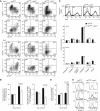
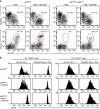
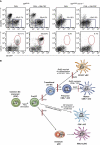
Dendritic cells (DCs) have critical roles in the induction of the adaptive immune response. The transcription factors Id2, Batf3 and Irf-8 are required for many aspects of murine DC differentiation including development of CD8α(+) and CD103(+) DCs. How they regulate DC subset specification is not completely understood. Using an Id2-GFP reporter system, we show that Id2 is broadly expressed in all cDC subsets with the highest expression in CD103(+) and CD8α(+) lineages. Notably, CD103(+) DCs were the only DC able to constitutively cross-present cell-associated antigens in vitro. Irf-8 deficiency affected loss of development of virtually all conventional DCs (cDCs) while Batf3 deficiency resulted in the development of Sirp-α(-) DCs that had impaired survival. Exposure to GM-CSF during differentiation induced expression of CD103 in Id2-GFP(+) DCs. It did not restore cross-presenting capacity to Batf3(-/-) or CD103(-)Sirp-α(-)DCs in vitro. Thus, Irf-8 and Batf3 regulate distinct stages in DC differentiation during the development of cDCs. Genetic mapping DC subset differentiation using Id2-GFP may have broad implications in understanding the interplay of DC subsets during protective and pathological immune responses.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, Sher A (2003) Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood 101: 305–310 - PubMed
-
- Allan RS, Smith CM, Belz GT, van Lint AL, Wakim LM, Heath WR, Carbone FR (2003) Epidermal viral immunity induced by CD8alpha+ dendritic cells but not by Langerhans cells. Science 301: 1925–1928 - PubMed
-
- Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, Salama A, Movassaghi K, Opitz C, Mages HW, Henn V, Kloetzel PM, Gurka S, Kroczek RA (2010) Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med 207: 1273–1281 - PMC - PubMed
-
- Barnden MJ, Allison J, Heath WR, Carbone FR (1998) Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76: 34–40 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

