Datamining approach for automation of diagnosis of breast cancer in immunohistochemically stained tissue microarray images
- PMID: 21589855
- PMCID: PMC3095117
- DOI: 10.2174/1874431101004010086
Datamining approach for automation of diagnosis of breast cancer in immunohistochemically stained tissue microarray images
Abstract
Cancer of the breast is the second most common human neoplasm, accounting for approximately one quarter of all cancers in females after cervical carcinoma. Estrogen receptor (ER), Progesteron receptor and human epidermal growth factor receptor (HER-2/neu) expressions play an important role in diagnosis and prognosis of breast carcinoma. Tissue microarray (TMA) technique is a high throughput technique which provides a standardized set of images which are uniformly stained, facilitating effective automation of the evaluation of the specimen images. TMA technique is widely used to evaluate hormone expression for diagnosis of breast cancer. If one considers the time taken for each of the steps in the tissue microarray process workflow, it can be observed that the maximum amount of time is taken by the analysis step. Hence, automated analysis will significantly reduce the overall time required to complete the study. Many tools are available for automated digital acquisition of images of the spots from the microarray slide. Each of these images needs to be evaluated by a pathologist to assign a score based on the staining intensity to represent the hormone expression, to classify them into negative or positive cases. Our work aims to develop a system for automated evaluation of sets of images generated through tissue microarray technique, representing the ER expression images and HER-2/neu expression images. Our study is based on the Tissue Microarray Database portal of Stanford university at http://tma.stanford.edu/cgi-bin/cx?n=her1, which has made huge number of images available to researchers. We used 171 images corresponding to ER expression and 214 images corresponding to HER-2/neu expression of breast carcinoma. Out of the 171 images corresponding to ER expression, 104 were negative and 67 were representing positive cases. Out of the 214 images corresponding to HER-2/neu expression, 112 were negative and 102 were representing positive cases. Our method has 92.31% sensitivity and 93.18% specificity for ER expression image classification and 96.67% sensitivity and 88.24% specificity for HER-2/neu expression image classification.
Keywords: Automated breast cancer diagnosis; Decision support systems; Image analysis; Tissue microarray; staining expression quantification..
Figures


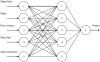
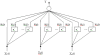
References
-
- Website of University of Pittsburgh Cancer Institute, Tissue MicroArray (TMA) Laboratory. Available from: http://www.upci. upmc.edu/tarps/index.cfm . [Accessed on 25.08.2009].
-
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, et al. Tissue microarray for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. - PubMed
-
- Shergill SI, Arya M. Tissue microarrays. Expert Rev Mol Diagn. 2004;4(4):421–3. - PubMed
-
- Venkataraman G, Ananthanaranayanan V. Tissue microarrays: Potential in the Indian subcontinent. Indian J Cancer. 2005;42(1):9–14. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
