Structural similarity and classification of protein interaction interfaces
- PMID: 21589874
- PMCID: PMC3093400
- DOI: 10.1371/journal.pone.0019554
Structural similarity and classification of protein interaction interfaces
Abstract
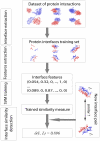
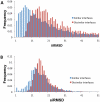
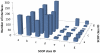
Interactions between proteins play a key role in many cellular processes. Studying protein-protein interactions that share similar interaction interfaces may shed light on their evolution and could be helpful in elucidating the mechanisms behind stability and dynamics of the protein complexes. When two complexes share structurally similar subunits, the similarity of the interaction interfaces can be found through a structural superposition of the subunits. However, an accurate detection of similarity between the protein complexes containing subunits of unrelated structure remains an open problem. Here, we present an alignment-free machine learning approach to measure interface similarity. The approach relies on the feature-based representation of protein interfaces and does not depend on the superposition of the interacting subunit pairs. Specifically, we develop an SVM classifier of similar and dissimilar interfaces and derive a feature-based interface similarity measure. Next, the similarity measure is applied to a set of 2,806×2,806 binary complex pairs to build a hierarchical classification of protein-protein interactions. Finally, we explore case studies of similar interfaces from each level of the hierarchy, considering cases when the subunits forming interactions are either homologous or structurally unrelated. The analysis has suggested that the positions of charged residues in the homologous interfaces are not necessarily conserved and may exhibit more complex conservation patterns.
Conflict of interest statement
Figures




References
-
- Alberts B. Essential cell biology : an introduction to the molecular biology of the cell. New York: Garland Pub; 1998. p. 1 v. (various pagings).
-
- Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. - PubMed
-
- Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, et al. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry. 2005;44:7035–7047. - PMC - PubMed
-
- Abbasi I, Githure J, Ochola JJ, Agure R, Koech DK, et al. Diagnosis of Wuchereria bancrofti infection by the polymerase chain reaction employing patients' sputum. Parasitol Res. 1999;85:844–849. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

