Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex
- PMID: 21589891
- PMCID: PMC3093354
- DOI: 10.1371/journal.pgen.1002074
Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex
Abstract
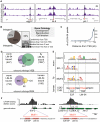
DRM is a conserved transcription factor complex that includes E2F/DP and pRB family proteins and plays important roles in development and cancer. Here we describe new aspects of DRM binding and function revealed through genome-wide analyses of the Caenorhabditis elegans DRM subunit LIN-54. We show that LIN-54 DNA-binding activity recruits DRM to promoters enriched for adjacent putative E2F/DP and LIN-54 binding sites, suggesting that these two DNA-binding moieties together direct DRM to its target genes. Chromatin immunoprecipitation and gene expression profiling reveals conserved roles for DRM in regulating genes involved in cell division, development, and reproduction. We find that LIN-54 promotes expression of reproduction genes in the germline, but prevents ectopic activation of germline-specific genes in embryonic soma. Strikingly, C. elegans DRM does not act uniformly throughout the genome: the DRM recruitment motif, DRM binding, and DRM-regulated embryonic genes are all under-represented on the X chromosome. However, germline genes down-regulated in lin-54 mutants are over-represented on the X chromosome. We discuss models for how loss of autosome-bound DRM may enhance germline X chromosome silencing. We propose that autosome-enriched binding of DRM arose in C. elegans as a consequence of germline X chromosome silencing and the evolutionary redistribution of germline-expressed and essential target genes to autosomes. Sex chromosome gene regulation may thus have profound evolutionary effects on genome organization and transcriptional regulatory networks.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Opposing activities of DRM and MES-4 tune gene expression and X-chromosome repression in Caenorhabditis elegans germ cells.G3 (Bethesda). 2014 Jan 10;4(1):143-53. doi: 10.1534/g3.113.007849. G3 (Bethesda). 2014. PMID: 24281426 Free PMC article.
-
Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB).Development. 2006 Aug;133(16):3147-57. doi: 10.1242/dev.02490. Epub 2006 Jul 19. Development. 2006. PMID: 16854972
-
DREAM interrupted: severing LIN-35-MuvB association in Caenorhabditis elegans impairs DREAM function but not its chromatin localization.Genetics. 2022 Jul 4;221(3):iyac073. doi: 10.1093/genetics/iyac073. Genetics. 2022. PMID: 35554539 Free PMC article.
-
Regulation of the X chromosomes in Caenorhabditis elegans.Cold Spring Harb Perspect Biol. 2014 Mar 1;6(3):a018366. doi: 10.1101/cshperspect.a018366. Cold Spring Harb Perspect Biol. 2014. PMID: 24591522 Free PMC article. Review.
-
LIN-35 beyond its classical roles: its function in the stress response.Int J Dev Biol. 2021;65(4-5-6):377-382. doi: 10.1387/ijdb.200194rn. Int J Dev Biol. 2021. PMID: 32930365 Review.
Cited by
-
Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes.PLoS Genet. 2012;8(3):e1002542. doi: 10.1371/journal.pgen.1002542. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412383 Free PMC article.
-
E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response.Genes Dev. 2016 Sep 15;30(18):2062-2075. doi: 10.1101/gad.283317.116. Epub 2016 Sep 29. Genes Dev. 2016. PMID: 27688402 Free PMC article.
-
Chlamydomonas CHT7 Is Required for an Effective Quiescent State by Regulating Nutrient-Responsive Cell Cycle Gene Expression.Plant Cell. 2020 Apr;32(4):1240-1269. doi: 10.1105/tpc.19.00628. Epub 2020 Jan 30. Plant Cell. 2020. PMID: 32001503 Free PMC article.
-
The DREAM complex promotes gene body H2A.Z for target repression.Genes Dev. 2015 Mar 1;29(5):495-500. doi: 10.1101/gad.255810.114. Genes Dev. 2015. PMID: 25737279 Free PMC article.
-
DREAM represses distinct targets by cooperating with different THAP domain proteins.Cell Rep. 2021 Oct 19;37(3):109835. doi: 10.1016/j.celrep.2021.109835. Cell Rep. 2021. PMID: 34686342 Free PMC article.
References
-
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. - PubMed
-
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. - PubMed
-
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

