Phylogeography and genetic structuring of European nine-spined sticklebacks (Pungitius pungitius)-mitochondrial DNA evidence
- PMID: 21589917
- PMCID: PMC3092751
- DOI: 10.1371/journal.pone.0019476
Phylogeography and genetic structuring of European nine-spined sticklebacks (Pungitius pungitius)-mitochondrial DNA evidence
Abstract
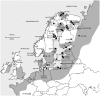
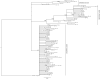
As a consequence of colonisation from different glacial refugia, many northern European taxa are split into distinct western and eastern lineages. However, as for the nine-spined stickleback (Pungitius pungitius), the exact location of the contact zone between lineages often remains poorly known. We assessed the genetic differentiation and diversity in the nine-spined stickleback within Europe using 1037 base pairs of cytochrome b sequence for 320 individuals from 57 locations, including pond, lake, river, and coastal habitats. Our main aims were (i) to locate the contact zone between the previously recognized western and eastern lineages, (ii) investigate latitudinal patterns in genetic diversity, (iii) compare genetic diversity among different habitat types, and (iv) date the known split between eastern and western lineages. The data revealed the split between eastern and western to be located across the Danish Straits and roughly following the Norway/Sweden border to the North. Reference sites from Canada form their own clades, and one of the Canadian sites was found to have a haplotype common to the Eastern European lineage, possibly representing an ancestral polymorphism. The split between the two European clades was dated to approximately 1.48 million years ago (Mya), and between Canada and Europe to approximately 1.62 Mya. After controlling for habitat effects, nucleotide (but not haplotype) diversity across populations decreased with increasing latitude. Coastal populations showed significantly higher haplotype diversity (but not nucleotide diversity) than pond populations, but there were no detectable differences in haplotype diversity among different freshwater habitat types (viz. river, lake and pond populations), or between coastal and lake/river populations. Sequences were found to cluster according to their geographic proximity, rather than by habitat type, and all habitat types were found within each major clade, implying that colonisation and adaptation between the coastal and freshwater environments in different regions must have occurred in parallel.
Conflict of interest statement
Figures

References
-
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. - PubMed
-
- Webb T, Bartlein P. Global changes during the last 3 million years: climatic controls and biotic response. Annu Rev Ecol Syst. 1992;23:141–173.
-
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998;7:453–464. - PubMed
-
- Bernatchez L, Osinov A. Genetic diversity of trout (genus Salmo) from its most eastern native range based on mitochondrial DNA and nuclear gene variation. Mol Ecol. 1995;4:285–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

