Phylogenetic and similarity analysis of HTLV-1 isolates from HIV-coinfected patients from the south and southeast regions of Brazil
- PMID: 21591992
- PMCID: PMC3252815
- DOI: 10.1089/AID.2011.0117
Phylogenetic and similarity analysis of HTLV-1 isolates from HIV-coinfected patients from the south and southeast regions of Brazil
Abstract
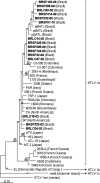
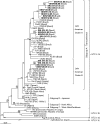
HTLV-1 is endemic in Brazil and HIV/HTLV-1 coinfection has been detected, mostly in the northeast region. Cosmopolitan HTLV-1a is the main subtype that circulates in Brazil. This study characterized 17 HTLV-1 isolates from HIV coinfected patients of southern (n=7) and southeastern (n=10) Brazil. HTLV-1 provirus DNA was amplified by nested PCR (env and LTR) and sequenced. Env sequences (705 bp) from 15 isolates and LTR sequences (731 bp) from 17 isolates showed 99.5% and 98.8% similarity among sequences, respectively. Comparing these sequences with ATK (HTLV-1a) and Mel5 (HTLV-1c) prototypes, similarities of 99% and 97.4%, respectively, for env and LTR with ATK, and 91.6% and 90.3% with Mel5, were detected. Phylogenetic analysis showed that all sequences belonged to the transcontinental subgroup A of the Cosmopolitan subtype, clustering in two Latin American clusters.
Figures
References
-
- Proietti FA. Carneiro-Proietti ABF. Catalan-Soares BC. Murphy EL. Global epidemiology of HTLV-1 infection and associated diseases. Oncogene. 2005;24:6058–6068. - PubMed
-
- Carneiro-Proietti AB. Catalan-Soares BC. Proietti FA. GIPH (Interdisciplinary HTLV-I/IIResearch Group): Human T cell lymphotropic viruses (HTLV-I/II) in South America: Should it be a public health concern? J Biomed Sci. 2002;9:587–595. - PubMed
-
- Alcantara LC. de Oliveira T. Gordon M, et al. Tracing the origin of Brazilian HTLV-1 as determined by analysis of host and viral genes. AIDS. 2006;20(5):780–782. - PubMed
-
- Ratner L. Philpott T. Trowbridge DB. Nucleotide sequence analysis of isolates of human T-lymphotropic virus type I of diverse geographical origins. AIDS Res Hum Retroviruses. 1991;7(11):923–941. - PubMed
-
- Yamashita M. Veronesi R. Menna-Barreto M, et al. Molecular epidemiology of human T-cell leukemia virus type I (HTLV-1) Brazil: The predominant HTLV-1s in South America differ from HTLV-l of Japan and Africa, as well as those of Japanese immigrants and their relatives in Brazil. Virology. 1999;261(1):59–69. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

