Optimal tumor sampling for immunostaining of biomarkers in breast carcinoma
- PMID: 21592345
- PMCID: PMC3218938
- DOI: 10.1186/bcr2882
Optimal tumor sampling for immunostaining of biomarkers in breast carcinoma
Abstract
Introduction: Biomarkers, such as Estrogen Receptor, are used to determine therapy and prognosis in breast carcinoma. Immunostaining assays of biomarker expression have a high rate of inaccuracy; for example, estimates are as high as 20% for Estrogen Receptor. Biomarkers have been shown to be heterogeneously expressed in breast tumors and this heterogeneity may contribute to the inaccuracy of immunostaining assays. Currently, no evidence-based standards exist for the amount of tumor that must be sampled in order to correct for biomarker heterogeneity. The aim of this study was to determine the optimal number of 20X fields that are necessary to estimate a representative measurement of expression in a whole tissue section for selected biomarkers: ER, HER-2, AKT, ERK, S6K1, GAPDH, Cytokeratin, and MAP-Tau.
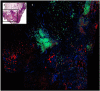
Methods: Two collections of whole tissue sections of breast carcinoma were immunostained for biomarkers. Expression was quantified using the Automated Quantitative Analysis (AQUA) method of quantitative immunofluorescence. Simulated sampling of various numbers of fields (ranging from one to thirty five) was performed for each marker. The optimal number was selected for each marker via resampling techniques and minimization of prediction error over an independent test set.
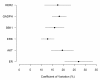
Results: The optimal number of 20X fields varied by biomarker, ranging between three to fourteen fields. More heterogeneous markers, such as MAP-Tau protein, required a larger sample of 20X fields to produce representative measurement.
Conclusions: The optimal number of 20X fields that must be sampled to produce a representative measurement of biomarker expression varies by marker with more heterogeneous markers requiring a larger number. The clinical implication of these findings is that breast biopsies consisting of a small number of fields may be inadequate to represent whole tumor biomarker expression for many markers. Additionally, for biomarkers newly introduced into clinical use, especially if therapeutic response is dictated by level of expression, the optimal size of tissue sample must be determined on a marker-by-marker basis.
Figures

References
-
- Ross J, Symmans W, Pusztai L, Hortobagyi G. Breast cancer biomarkers. Advances in Clinical Chemistry. 2005;40:99–125. - PubMed
-
- Hammond M, Hayes D, Dowsett M, Allred D, Hagerty K, Badve S, Fitzgibbons P, Francis G, Goldstein N, Hayes M, Hicks D, Lester S, Love R, Mangu P, McShane L, Miller K, Osborne C, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz J, Sweep F, Taube S, Torlakovic E, Valenstein P, Viale G, Visscher D, Wheeler T, Williams R. et al. American society of clinical oncology/college of American pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Journal of Clinical Oncology. 2010;28:2784. doi: 10.1200/JCO.2009.25.6529. - DOI - PMC - PubMed
-
- Vance G, Barry T, Bloom K, Fitzgibbons P, Hicks D, Jenkins R, Persons D, Tubbs R, Hammond M. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Archives of Pathology & Laboratory Medicine. 2009;133:611–612. - PubMed
-
- Nassar A, Radhakrishnan A, Cabrero I, Cotsonis G, Cohen C. Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Applied Immunohistochemistry & Molecular Morphology. 2010. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

