Causal graph-based analysis of genome-wide association data in rheumatoid arthritis
- PMID: 21592391
- PMCID: PMC3118953
- DOI: 10.1186/1745-6150-6-25
Causal graph-based analysis of genome-wide association data in rheumatoid arthritis
Abstract
Background: GWAS owe their popularity to the expectation that they will make a major impact on diagnosis, prognosis and management of disease by uncovering genetics underlying clinical phenotypes. The dominant paradigm in GWAS data analysis so far consists of extensive reliance on methods that emphasize contribution of individual SNPs to statistical association with phenotypes. Multivariate methods, however, can extract more information by considering associations of multiple SNPs simultaneously. Recent advances in other genomics domains pinpoint multivariate causal graph-based inference as a promising principled analysis framework for high-throughput data. Designed to discover biomarkers in the local causal pathway of the phenotype, these methods lead to accurate and highly parsimonious multivariate predictive models. In this paper, we investigate the applicability of causal graph-based method TIE* to analysis of GWAS data. To test the utility of TIE*, we focus on anti-CCP positive rheumatoid arthritis (RA) GWAS datasets, where there is a general consensus in the community about the major genetic determinants of the disease.
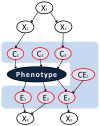
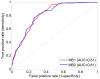
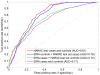
Results: Application of TIE* to the North American Rheumatoid Arthritis Cohort (NARAC) GWAS data results in six SNPs, mostly from the MHC locus. Using these SNPs we develop two predictive models that can classify cases and disease-free controls with an accuracy of 0.81 area under the ROC curve, as verified in independent testing data from the same cohort. The predictive performance of these models generalizes reasonably well to Swedish subjects from the closely related but not identical Epidemiological Investigation of Rheumatoid Arthritis (EIRA) cohort with 0.71-0.78 area under the ROC curve. Moreover, the SNPs identified by the TIE* method render many other previously known SNP associations conditionally independent of the phenotype.
Conclusions: Our experiments demonstrate that application of TIE* captures maximum amount of genetic information about RA in the data and recapitulates the major consensus findings about the genetic factors of this disease. In addition, TIE* yields reproducible markers and signatures of RA. This suggests that principled multivariate causal and predictive framework for GWAS analysis empowers the community with a new tool for high-quality and more efficient discovery.
Reviewers: This article was reviewed by Prof. Anthony Almudevar, Dr. Eugene V. Koonin, and Prof. Marianthi Markatou.
Figures


References
-
- Chang M, Rowland CM, Garcia VE, Schrodi SJ, Catanese JJ, van der Helm-van Mil AH, Ardlie KG, Amos CI, Criswell LA, Kastner DL, Gregersen PK, Kurreeman FA, Toes RE, Huizinga TW, Seldin MF, Begovich AB. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4:e1000107. - PMC - PubMed
-
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. - PMC - PubMed
-
- Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, Chang M, Catanese JJ, Ding B, Wong S, van der Helm-van Mil AH, Neale BM, Coblyn J, Cui J, Tak PP, Wolbink GJ, Crusius JB, van der Horst-Bruinsma IE, Criswell LA, Amos CI, Seldin MF, Kastner DL, Ardlie KG, Alfredsson L, Costenbader KH, Altshuler D, Huizinga TW, Shadick NA, Weinblatt ME, de VN, Worthington J, Seielstad M, Toes RE, Karlson EW, Begovich AB, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

