Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotiana benthamiana and their correlation with the levels of developmental miRNAs
- PMID: 21592402
- PMCID: PMC3166278
- DOI: 10.1186/1743-422X-8-238
Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotiana benthamiana and their correlation with the levels of developmental miRNAs
Abstract
Background: Whitefly-transmitted geminiviruses (begomoviruses) are a major limiting factor for the production of numerous dicotyledonous crops throughout the world. Begomoviruses differ in the number of components that make up their genomes and association with satellites, and yet they cause strikingly similar phenotypes, such as leaf curling, chlorosis and stunted plant growth. MicroRNAs (miRNAs) are small endogenous RNAs that regulate plant growth and development. The study described here was aimed at investigating the effects of each virus encoded gene on the levels of developmental miRNAs to identify common trends between distinct begomoviruses.
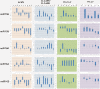
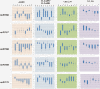
Results: All genes encoded by four distinct begomoviruses (African cassava mosaic virus [ACMV], Cabbage leaf curl virus [CbLCuV], Tomato yellow leaf curl virus [TYLCV] and Cotton leaf curl virus/Cotton leaf curl betasatellite [CLCuV/CLCuMB]) were expressed from a Potato virus X (PVX) vector in Nicotiana benthamiana. Changes in the levels of ten miRNAs in response to the virus genes were determined by northern blotting using specific miRNA probes. For the monopartite begomoviruses (TYLCV and CLCuMV) the V2 gene product was identified as the major symptom determinant while for bipartite begomoviruses (ACMV and CbLCuV) more than one gene appears to contribute to symptoms and this is reflected in changes in miRNA levels. The phenotype induced by expression of the βC1 gene of the betasatellite CLCuMB was the most distinct and consisted of leaf curling, vein swelling, thick green veins and enations and the pattern of changes in miRNA levels was the most distinct.
Conclusions: Our results have identified symptom determinants encoded by begomoviruses and show that developmental abnormalities caused by transient expression of begomovirus genes correlates with altered levels of developmental miRNAs. Additionally, all begomovirus genes were shown to modulate miRNA levels, the first time this has been shown to be the case.
Figures









References
-
- Zaitlin M, Hull R. Plant Virus-Host Interaction. Ann Rev Plant Physiol. 1987;38:291–315. doi: 10.1146/annurev.pp.38.060187.001451. - DOI
-
- Voinnet O. Non-cell autonomous RNA silencing: insights from viral infections. FEBS Lett. 2005. pp. 579–5858. - PubMed
-
- Navarro B, Pantaleo V, Gisel A, Moxon S, Dalmay T, Bisztray G, Di F Serio, Burgyan J. Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS One. 2009;4:e7686. doi: 10.1371/journal.pone.0007686. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

