Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes
- PMID: 21592595
- PMCID: PMC3116240
- DOI: 10.1016/j.tins.2011.04.001
Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes
Abstract
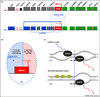
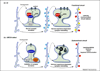
Angelman syndrome (AS) is a severe genetic disorder caused by mutations or deletions of the maternally inherited UBE3A gene. UBE3A encodes an E3 ubiquitin ligase that is expressed biallelically in most tissues but is maternally expressed in almost all neurons. In this review, we describe recent advances in understanding the expression and function of UBE3A in the brain and the etiology of AS. We highlight current AS model systems, epigenetic mechanisms of UBE3A regulation, and the identification of potential UBE3A substrates in the brain. In the process, we identify major gaps in our knowledge that, if bridged, could move us closer to identifying treatments for this debilitating neurodevelopmental disorder.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Steffenburg S, et al. Autism in Angelman syndrome: a population-based study. Pediatr Neurol. 1996;14(2):131–136. - PubMed
-
- Williams CA, et al. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140(5):413–418. - PubMed
-
- Peters SU, et al. Autism in Angelman syndrome: implications for autism research. Clin Genet. 2004;66(6):530–536. - PubMed
-
- Trillingsgaard A, JR OS. Autism in Angelman syndrome: an exploration of comorbidity. Autism. 2004;8(2):163–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

