Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity
- PMID: 21593585
- PMCID: PMC3127101
- DOI: 10.4161/rna.8.2.15013
Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity
Abstract
In order to survive and propagate, RNA viruses must achieve a balance between the capacity for adaptation to new environmental conditions or host cells with the need to maintain an intact and replication competent genome. Several virus families in the order Nidovirales, such as the coronaviruses (CoVs) must achieve these objectives with the largest and most complex replicating RNA genomes known, up to 32 kb of positive-sense RNA. The CoVs encode sixteen nonstructural proteins (nsp 1-16) with known or predicted RNA synthesis and modification activities, and it has been proposed that they are also responsible for the evolution of large genomes. The CoVs, including murine hepatitis virus (MHV) and SARS-CoV, encode a 3'-to-5' exoribonuclease activity (ExoN) in nsp14. Genetic inactivation of ExoN activity in engineered SARS-CoV and MHV genomes by alanine substitution at conserved DE-D-D active site residues results in viable mutants that demonstrate 15- to 20-fold increases in mutation rates, up to 18 times greater than those tolerated for fidelity mutants of other RNA viruses. Thus nsp14-ExoN is essential for replication fidelity, and likely serves either as a direct mediator or regulator of a more complex RNA proofreading machine, a process previously unprecedented in RNA virus biology. Elucidation of the mechanisms of nsp14-mediated proofreading will have major implications for our understanding of the evolution of RNA viruses, and also will provide a robust model to investigate the balance between fidelity, diversity and pathogenesis. The discovery of a protein distinct from a viral RdRp that regulates replication fidelity also raises the possibility that RNA genome replication fidelity may be adaptable to differing replication environments and selective pressures, rather than being a fixed determinant.
Figures



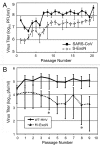
References
-
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
