CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer
- PMID: 21593597
- PMCID: PMC3121973
- DOI: 10.4161/epi.6.5.15667
CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer
Abstract
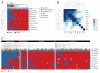
Triple-negative breast cancer (TNBC) occurs in approximately 15% of all breast cancer patients, and the incidence of TNBC is greatly increased in BRCA1 mutation carriers. This study aimed to assess the impact of BRCA1 promoter methylation with respect to breast cancer subtypes in sporadic disease. Tissue microarrays (TMAs) were constructed representing tumors from 303 patients previously screened for BRCA1 germline mutations, of which a subset of 111 sporadic tumors had previously been analyzed with respect to BRCA1 methylation. Additionally, a set of eight tumors from BRCA1 mutation carriers were included on the TMAs. Expression analysis was performed on TMAs by immunohistochemistry (IHC) for BRCA1, pRb, p16, p53, PTEN, ER, PR, HER2, CK5/6, EGFR, MUC1 and Ki-67. Data on BRCA1 aberrations and IHC expression was examined with respect to breast cancer-specific survival. The results demonstrate that CpG island hypermethylation of BRCA1 significantly associates with the basal/triple-negative subtype. Low expression of pRb, and high/intense p16, were associated with BRCA1 promoter hypermethylation, and the same effects were seen in BRCA1 mutated tumors. The expression patterns of BRCA1, pRb, p16 and PTEN were highly correlated, and define a subgroup of TNBCs characterized by BRCA1 aberrations, high Ki-67 (≥ 40%) and favorable disease outcome. In conclusion, our findings demonstrate that epigenetic inactivation of the BRCA1 gene associates with RB/p16 dysfunction in promoting TNBCs. The clinical implications relate to the potential use of targeted treatment based on PARP inhibitors in sporadic TNBCs, wherein CpG island hypermethylation of BRCA1 represents a potential marker of therapeutic response.
Figures


References
-
- Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. - PubMed
-
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
