Overexpression of human virus surface glycoprotein precursors induces cytosolic unfolded protein response in Saccharomyces cerevisiae
- PMID: 21595909
- PMCID: PMC3120639
- DOI: 10.1186/1475-2859-10-37
Overexpression of human virus surface glycoprotein precursors induces cytosolic unfolded protein response in Saccharomyces cerevisiae
Abstract
Background: The expression of human virus surface proteins, as well as other mammalian glycoproteins, is much more efficient in cells of higher eukaryotes rather than yeasts. The limitations to high-level expression of active viral surface glycoproteins in yeast are not well understood. To identify possible bottlenecks we performed a detailed study on overexpression of recombinant mumps hemagglutinin-neuraminidase (MuHN) and measles hemagglutinin (MeH) in yeast Saccharomyces cerevisiae, combining the analysis of recombinant proteins with a proteomic approach.
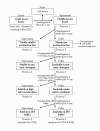
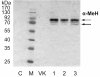
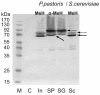
Results: Overexpressed recombinant MuHN and MeH proteins were present in large aggregates, were inactive and totally insoluble under native conditions. Moreover, the majority of recombinant protein was found in immature form of non-glycosylated precursors. Fractionation of yeast lysates revealed that the core of viral surface protein aggregates consists of MuHN or MeH disulfide-linked multimers involving eukaryotic translation elongation factor 1A (eEF1A) and is closely associated with small heat shock proteins (sHsps) that can be removed only under denaturing conditions. Complexes of large Hsps seem to be bound to aggregate core peripherally as they can be easily removed at high salt concentrations. Proteomic analysis revealed that the accumulation of unglycosylated viral protein precursors results in specific cytosolic unfolded protein response (UPR-Cyto) in yeast cells, characterized by different action and regulation of small Hsps versus large chaperones of Hsp70, Hsp90 and Hsp110 families. In contrast to most environmental stresses, in the response to synthesis of recombinant MuHN and MeH, only the large Hsps were upregulated whereas sHsps were not. Interestingly, the amount of eEF1A was also increased during this stress response.
Conclusions: Inefficient translocation of MuHN and MeH precursors through ER membrane is a bottleneck for high-level expression in yeast. Overexpression of these recombinant proteins induces the UPR's cytosolic counterpart, the UPR-Cyto, which represent a subset of proteins involved in the heat-shock response. The involvement of eEF1A may explain the mechanism by which only large chaperones, but not small Hsps are upregulated during this stress response. Our study highlights important differences between viral surface protein expression in yeast and mammalian cells at the first stage of secretory pathway.
Figures
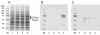



References
-
- Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C, Porro D, Ferrer P, Tutino ML, Mattanovich D, Villaverde A. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

