Mitochondrial RNA processing in trypanosomes
- PMID: 21596134
- PMCID: PMC3148333
- DOI: 10.1016/j.resmic.2011.04.015
Mitochondrial RNA processing in trypanosomes
Abstract
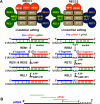
The mitochondrial genome of trypanosomes is composed of ∼50 maxicircles and thousands of minicircles. Maxi-(∼25 kb) and mini-(∼1 kb)circles are catenated and packed into a dense structure called a kinetoplast. Both types of circular DNA are transcribed by a phage-like RNA polymerase: maxicircles yield multicistronic rRNA and mRNA precursors, while guide RNA (gRNA) precursors are produced from minicircles. To function in mitochondrial translation, pre-mRNAs must undergo a nucleolytic processing and 3' modifications, and often uridine insertion/deletion editing. gRNAs, which represent short (50-60 nt) RNAs directing editing reactions, are produced by 3' nucleolytic processing of a much longer precursor followed by 3' uridylation. Ribosomal RNAs are excised from precursors and their 3' ends are also trimmed and uridylated. All tRNAs are imported from the cytoplasm and some are further modified and edited in the mitochondrial matrix. Historically, the fascinating phenomenon of RNA editing has been extensively studied as an isolated pathway in which nuclear-encoded proteins mediate interactions of maxi- and minicircle transcripts to create open reading frames. However, recent studies unraveled a highly integrated network of mitochondrial genome expression including critical pre- and post-editing 3' mRNA processing, and gRNA and rRNA maturation steps. Here we focus on RNA 3' adenylation and uridylation as processes essential for biogenesis, stability and functioning of mitochondrial RNAs.
Copyright © 2011 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved.
Figures



References
-
- Alfonzo JD, Thiemann OH, Simpson L. Purification and characterization of MAR1 - A mitochondrial associated ribonuclease from Leishmania tarentolae. J. Biol. Chem. 1998;273:30003–30011. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

