Resolution of gene regulatory conflicts caused by combinations of antibiotics
- PMID: 21596308
- PMCID: PMC3143497
- DOI: 10.1016/j.molcel.2011.04.016
Resolution of gene regulatory conflicts caused by combinations of antibiotics
Abstract
Regulatory conflicts occur when two signals that individually trigger opposite cellular responses are present simultaneously. Here, we investigate regulatory conflicts in the bacterial response to antibiotic combinations. We use an Escherichia coli promoter-GFP library to study the transcriptional response of many promoters to either additive or antagonistic drug pairs at fine two-dimensional (2D) resolution of drug concentration. Surprisingly, we find that this data set can be characterized as a linear sum of only two principal components. Component one, accounting for over 70% of the response, represents the response to growth inhibition by the drugs. Component two describes how regulatory conflicts are resolved. For the additive drug pair, conflicts are resolved by linearly interpolating the single drug responses, while for the antagonistic drug pair, the growth-limiting drug dominates the response. Importantly, for a given drug pair, the same conflict resolution strategy applies to almost all genes. These results provide a recipe for predicting gene expression responses to antibiotic combinations.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
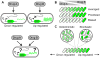
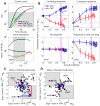



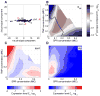
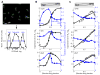
Comment in
-
Mixed messages: how bacteria resolve conflicting signals.Mol Cell. 2011 May 20;42(4):405-6. doi: 10.1016/j.molcel.2011.05.005. Mol Cell. 2011. PMID: 21596304 Free PMC article.
References
-
- Alberts B. Molecular biology of the cell. 5. New York: Abingdon, Garland Science; 2008.
-
- Bliss CI. The toxicity of poisons applied jointly. Annals of Applied Biology. 1939;26:585–615.
-
- Brazas MD, Hancock RE. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov Today. 2005;10:1245–1252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

