Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter
- PMID: 21596311
- PMCID: PMC3119483
- DOI: 10.1016/j.molcel.2011.03.028
Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter
Abstract
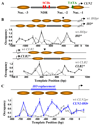
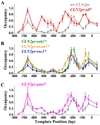
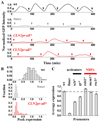
Nucleosome-depleted regions (NDRs) are ubiquitous on eukaryotic promoters. The formation of many NDRs cannot be readily explained by previously proposed mechanisms. Here, we carry out a focused study on a physiologically important NDR in the yeast CLN2 promoter (CLN2pr). We show that this NDR does not result from intrinsically unfavorable histone-DNA interaction. Instead, we identified eight conserved factor binding sites, including that of Reb1, Mcm1, and Rsc3, that cause the local nucleosome depletion. These nucleosome-depleting factors (NDFs) work redundantly, and simultaneously mutating all their binding sites eliminates CLN2pr NDR. The loss of the NDR induces unreliable "on/off" expression in individual cell cycles, but in the presence of the NDR, NDFs have little direct effect on transcription. We present bioinformatic evidence that the formation of many NDRs across the genome involves multiple NDFs. Our findings also provide significant insight into the composition and spatial organization of functional promoters.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. - PubMed
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

