Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay
- PMID: 21596314
- PMCID: PMC3123134
- DOI: 10.1016/j.molcel.2011.04.018
Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay
Abstract
Nonsense-mediated decay (NMD) degrades both normal and aberrant transcripts harboring stop codons in particular contexts. Mutations that perturb NMD cause neurological disorders in humans, suggesting that NMD has roles in the brain. Here, we identify a brain-specific microRNA-miR-128-that represses NMD and thereby controls batteries of transcripts in neural cells. miR-128 represses NMD by targeting the RNA helicase UPF1 and the exon-junction complex core component MLN51. The ability of miR-128 to regulate NMD is a conserved response occurring in frogs, chickens, and mammals. miR-128 levels are dramatically increased in differentiating neuronal cells and during brain development, leading to repressed NMD and upregulation of mRNAs normally targeted for decay by NMD; overrepresented are those encoding proteins controlling neuron development and function. Together, these results suggest the existence of a conserved RNA circuit linking the microRNA and NMD pathways that induces cell type-specific transcripts during development.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
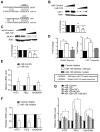

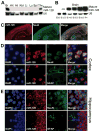
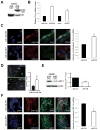
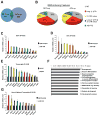
Comment in
-
Stay tuned: miRNA expression and nonsense-mediated decay in brain development.Mol Cell. 2011 May 20;42(4):407-8. doi: 10.1016/j.molcel.2011.05.002. Mol Cell. 2011. PMID: 21596305
Similar articles
-
A conserved microRNA/NMD regulatory circuit controls gene expression.RNA Biol. 2012 Jan;9(1):22-6. doi: 10.4161/rna.9.1.18010. Epub 2012 Jan 1. RNA Biol. 2012. PMID: 22258150 Free PMC article.
-
UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond.RNA. 2019 Apr;25(4):407-422. doi: 10.1261/rna.070136.118. Epub 2019 Jan 17. RNA. 2019. PMID: 30655309 Free PMC article. Review.
-
Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article.
-
A post-translational regulatory switch on UPF1 controls targeted mRNA degradation.Genes Dev. 2014 Sep 1;28(17):1900-16. doi: 10.1101/gad.245506.114. Genes Dev. 2014. PMID: 25184677 Free PMC article.
-
Defining nonsense-mediated mRNA decay intermediates in human cells.Methods. 2019 Feb 15;155:68-76. doi: 10.1016/j.ymeth.2018.12.005. Epub 2018 Dec 19. Methods. 2019. PMID: 30576707 Free PMC article. Review.
Cited by
-
Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact.Wiley Interdiscip Rev RNA. 2016 Sep;7(5):661-82. doi: 10.1002/wrna.1357. Epub 2016 May 13. Wiley Interdiscip Rev RNA. 2016. PMID: 27173476 Free PMC article. Review.
-
Progression of the pluripotent epiblast depends upon the NMD factor UPF2.Development. 2022 Nov 1;149(21):dev200764. doi: 10.1242/dev.200764. Epub 2022 Nov 7. Development. 2022. PMID: 36255229 Free PMC article.
-
An evolutionarily biased distribution of miRNA sites toward regulatory genes with high promoter-driven intrinsic transcriptional noise.BMC Evol Biol. 2014 Apr 4;14:74. doi: 10.1186/1471-2148-14-74. BMC Evol Biol. 2014. PMID: 24707827 Free PMC article.
-
Intron retention in viruses and cellular genes: Detention, border controls and passports.Wiley Interdiscip Rev RNA. 2018 May;9(3):e1470. doi: 10.1002/wrna.1470. Epub 2018 Mar 6. Wiley Interdiscip Rev RNA. 2018. PMID: 29508942 Free PMC article. Review.
-
MicroRNA profiling as tool for in vitro developmental neurotoxicity testing: the case of sodium valproate.PLoS One. 2014 Jun 4;9(6):e98892. doi: 10.1371/journal.pone.0098892. eCollection 2014. PLoS One. 2014. PMID: 24896083 Free PMC article.
References
-
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

