A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development
- PMID: 21596603
- PMCID: PMC3119599
- DOI: 10.1016/j.molcel.2011.04.020
A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development
Abstract
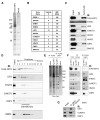
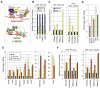
Epigenetic regulation of gene expression by histone-modifying corepressor complexes is central to normal animal development. The NAD(+)-dependent deacetylase and gene repressor SIRT1 removes histone H4K16 acetylation marks and facilitates heterochromatin formation. However, the mechanistic contribution of SIRT1 to epigenetic regulation at euchromatic loci and whether it acts in concert with other chromatin-modifying activities to control developmental gene expression programs remain unclear. We describe here a SIRT1 corepressor complex containing the histone H3K4 demethylase LSD1/KDM1A and several other LSD1-associated proteins. SIRT1 and LSD1 interact directly and play conserved and concerted roles in H4K16 deacetylation and H3K4 demethylation to repress genes regulated by the Notch signaling pathway. Mutations in Drosophila SIRT1 and LSD1 orthologs result in similar developmental phenotypes and genetically interact with the Notch pathway in Drosophila. These findings offer new insights into conserved mechanisms of epigenetic gene repression and regulation of development by SIRT1 in metazoans.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Comment in
-
A SirT'N repression for Notch.Mol Cell. 2011 Jun 10;42(5):559-60. doi: 10.1016/j.molcel.2011.05.017. Mol Cell. 2011. PMID: 21658598 Free PMC article.
References
-
- Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

