Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes
- PMID: 21596849
- PMCID: PMC3193274
- DOI: 10.1182/blood-2011-03-339705
Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes
Erratum in
- Blood. 2014 Jun 12;123(24):3843
Abstract
Inbred mice are a useful tool for studying the in vivo functions of platelets. Nonetheless, the mRNA signature of mouse platelets is not known. Here, we use paired-end next-generation RNA sequencing (RNA-seq) to characterize the polyadenylated transcriptomes of human and mouse platelets. We report that RNA-seq provides unprecedented resolution of mRNAs that are expressed across the entire human and mouse genomes. Transcript expression and abundance are often conserved between the 2 species. Several mRNAs, however, are differentially expressed in human and mouse platelets. Moreover, previously described functional disparities between mouse and human platelets are reflected in differences at the transcript level, including protease activated receptor-1, protease activated receptor-3, platelet activating factor receptor, and factor V. This suggests that RNA-seq is a useful tool for predicting differences in platelet function between mice and humans. Our next-generation sequencing analysis provides new insights into the human and murine platelet transcriptomes. The sequencing dataset will be useful in the design of mouse models of hemostasis and a catalyst for discovery of new functions of platelets. Access to the dataset is found in the "Introduction."
Figures

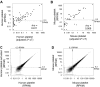
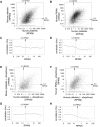

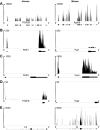

Comment in
-
A platelet transcriptome revolution.Blood. 2011 Oct 6;118(14):3760-1. doi: 10.1182/blood-2011-05-356600. Blood. 2011. PMID: 21980048 No abstract available.
References
-
- Jirouskova M, Shet AS, Johnson GJ. A guide to murine platelet structure, function, assays, and genetic alterations. J Thromb Haemost. 2007;5(4):661–669. - PubMed
-
- Tsakiris D, Scudder L, Hodivala-Dilke K, Hynes RO, Coller BS. Hemostasis in the mouse (Mus musculus): a review. Thromb Haemost. 1999;81(2):177–188. - PubMed
-
- Nieswandt B, Aktas B, Moers A, Sachs UJH. Platelets in atherothrombosis: lessons from mouse models. J Thromb Haemost. 2005;3(8):1725–1736. - PubMed
-
- Darrow AL, Fung-Leung WP, Ye RD, et al. Biological consequences of thrombin receptor deficiency in mice. Thromb Haemost. 1996;76(6):860–866. - PubMed
-
- Connolly AJ, Ishihara H, Kahn ML, Farese RV, Coughlin SR. Role of the thrombin receptor in development and evidence for a second receptor. Nature. 1996;381(6582):516–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

