Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice
- PMID: 21596949
- PMCID: PMC3135938
- DOI: 10.1104/pp.111.176016
Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice
Abstract
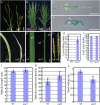
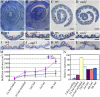
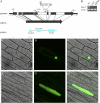
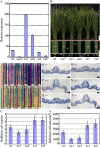
Leaf rolling is considered an important agronomic trait in rice (Oryza sativa) breeding. To understand the molecular mechanism controlling leaf rolling, we screened a rice T-DNA insertion population and isolated the outcurved leaf1 (oul1) mutant showing abaxial leaf rolling. The phenotypes were caused by knockout of Rice outermost cell-specific gene5 (Roc5), an ortholog of the Arabidopsis (Arabidopsis thaliana) homeodomain leucine zipper class IV gene GLABRA2. Interestingly, overexpression of Roc5 led to adaxially rolled leaves, whereas cosuppression of Roc5 resulted in abaxial leaf rolling. Bulliform cell number and size increased in oul1 and Roc5 cosuppression plants but were reduced in Roc5-overexpressing lines. The data indicate that Roc5 negatively regulates bulliform cell fate and development. Gene expression profiling, quantitative polymerase chain reaction, and RNA interference (RNAi) analyses revealed that Protodermal Factor Like (PFL) was probably down-regulated in oul1. The mRNA level of PFL was increased in Roc5-overexpressing lines, and PFL-RNAi transgenic plants exhibit reversely rolling leaves by reason of increases of bulliform cell number and size, indicating that Roc5 may have a conserved function. These are, to our knowledge, the first functional data for a gene encoding a homeodomain leucine zipper class IV transcriptional factor in rice that modulates leaf rolling.
Figures


References
-
- Abe M, Katsumata H, Komeda Y, Takahashi T. (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 - PubMed
-
- Abe M, Takahashi T, Komeda Y. (2001) Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J 26: 487–494 - PubMed
-
- Barrs HD, Weatherley PE. (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. J Biol Sci 15: 415–428
-
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y. (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225 - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

