A pharmacogenetic approach to identify mutant forms of α-galactosidase A that respond to a pharmacological chaperone for Fabry disease
- PMID: 21598360
- PMCID: PMC3170878
- DOI: 10.1002/humu.21530
A pharmacogenetic approach to identify mutant forms of α-galactosidase A that respond to a pharmacological chaperone for Fabry disease
Abstract
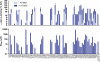
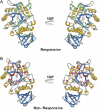
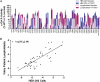
Fabry disease is caused by mutations in the gene (GLA) that encodes α-galactosidase A (α-Gal A). The iminosugar AT1001 (GR181413A, migalastat hydrochloride, 1-deoxygalactonojirimycin) is a pharmacological chaperone that selectively binds and stabilizes α-Gal A, increasing total cellular levels and activity for some mutant forms (defined as "responsive"). In this study, we developed a cell-based assay in cultured HEK-293 cells to identify mutant forms of α-Gal A that are responsive to AT1001. Concentration-dependent increases in α-Gal A activity in response to AT1001 were shown for 49 (60%) of 81 mutant forms. The responses of α-Gal A mutant forms were generally consistent with the responses observed in male Fabry patient-derived lymphoblasts. Importantly, the HEK-293 cell responses of 19 α-Gal A mutant forms to a clinically achievable concentration of AT1001 (10 µM) were generally consistent with observed increases in α-Gal A activity in peripheral blood mononuclear cells from male Fabry patients orally administered AT1001 during Phase 2 clinical studies. This indicates that the cell-based responses can identify mutant forms of α-Gal A that are likely to respond to AT1001 in vivo. Thus, the HEK-293 cell-based assay may be a useful aid in the identification of Fabry patients with AT1001-responsive mutant forms.
© 2011 Wiley-Liss, Inc.
Figures


References
-
- Asano N, Ishii S, Kizu H, Ikeda K, Yasuda K, Kato A, Martin OR, Fan JQ. In vitro inhibition and intracellular enhancement of lysosomal alpha-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur J Biochem. 2000;267:4179–4186. - PubMed
-
- Bekri S, Enica A, Ghafari T, Plaza G, Champenois I, Choukroun G, Unwin R, Jaeger P. Fabry disease in patients with end-stage renal failure: the potential benefits of screening. Nephron Clin Pract. 2005;101:c33–c38. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

