Apple skin patterning is associated with differential expression of MYB10
- PMID: 21599973
- PMCID: PMC3127826
- DOI: 10.1186/1471-2229-11-93
Apple skin patterning is associated with differential expression of MYB10
Abstract
Background: Some apple (Malus × domestica Borkh.) varieties have attractive striping patterns, a quality attribute that is important for determining apple fruit market acceptance. Most apple cultivars (e.g. 'Royal Gala') produce fruit with a defined fruit pigment pattern, but in the case of 'Honeycrisp' apple, trees can produce fruits of two different kinds: striped and blushed. The causes of this phenomenon are unknown.
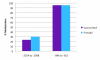
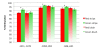
Results: Here we show that striped areas of 'Honeycrisp' and 'Royal Gala' are due to sectorial increases in anthocyanin concentration. Transcript levels of the major biosynthetic genes and MYB10, a transcription factor that upregulates apple anthocyanin production, correlated with increased anthocyanin concentration in stripes. However, nucleotide changes in the promoter and coding sequence of MYB10 do not correlate with skin pattern in 'Honeycrisp' and other cultivars differing in peel pigmentation patterns. A survey of methylation levels throughout the coding region of MYB10 and a 2.5 Kb region 5' of the ATG translation start site indicated that an area 900 bp long, starting 1400 bp upstream of the translation start site, is highly methylated. Cytosine methylation was present in all three contexts, with higher methylation levels observed for CHH and CHG (where H is A, C or T) than for CG. Comparisons of methylation levels of the MYB10 promoter in 'Honeycrisp' red and green stripes indicated that they correlate with peel phenotypes, with an enrichment of methylation observed in green stripes.
Conclusions: Differences in anthocyanin levels between red and green stripes can be explained by differential transcript accumulation of MYB10. Different levels of MYB10 transcript in red versus green stripes are inversely associated with methylation levels in the promoter region. Although observed methylation differences are modest, trends are consistent across years and differences are statistically significant. Methylation may be associated with the presence of a TRIM retrotransposon within the promoter region, but the presence of the TRIM element alone cannot explain the phenotypic variability observed in 'Honeycrisp'. We suggest that methylation in the MYB10 promoter is more variable in 'Honeycrisp' than in 'Royal Gala', leading to more variable color patterns in the peel of this cultivar.
Figures
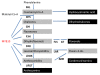


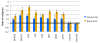


References
-
- Saure MC. External control of anthocyanin formation in apple: a review. Scientia Hort. 1990;42:181–218. doi: 10.1016/0304-4238(90)90082-P. - DOI
-
- Cliff M, Sandford K, Wismer W, Hampson C. Use of digital images for evaluation of factors responsible for visual preference of apples by consumers. HortScience. 2002;37:1127–1131.
-
- Eberhardt MV, Lee CY, Liu RH. Antioxidant activity of fresh apples. Nature. 2000;405:903–904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

