Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application
- PMID: 21602323
- PMCID: PMC3135926
- DOI: 10.1104/pp.111.175471
Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application
Abstract
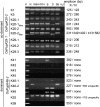
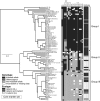
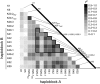
The major quantitative trait locus (QTL) Phosphorus uptake1 (Pup1) confers tolerance of phosphorus deficiency in soil and is currently one of the most promising QTLs for the development of tolerant rice (Oryza sativa) varieties. To facilitate targeted introgression of Pup1 into intolerant varieties, the gene models predicted in the Pup1 region in the donor variety Kasalath were used to develop gene-based molecular markers that are evenly distributed over the fine-mapped 278-kb QTL region. To validate the gene models and optimize the markers, gene expression analyses and partial allelic sequencing were conducted. The markers were tested in more than 80 diverse rice accessions revealing three main groups with different Pup1 allele constitution. Accessions with tolerant (group I) and intolerant (group III) Pup1 alleles were distinguished from genotypes with Kasalath alleles at some of the analyzed loci (partial Pup1; group II). A germplasm survey additionally confirmed earlier data showing that Pup1 is largely absent from irrigated rice varieties but conserved in varieties and breeding lines adapted to drought-prone environments. A core set of Pup1 markers has been defined, and sequence polymorphisms suitable for single-nucleotide polymorphism marker development for high-throughput genotyping were identified. Following a marker-assisted backcrossing approach, Pup1 was introgressed into two irrigated rice varieties and three Indonesian upland varieties. First phenotypic evaluations of the introgression lines suggest that Pup1 is effective in different genetic backgrounds and environments and that it has the potential to significantly enhance grain yield under field conditions.
Figures




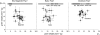
References
-
- Bernier J, Kumar A, Venuprasad R, Spaner D, Atlin G. (2007) A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47: 507–516
-
- Bernier J, Kumar A, Venuprasad R, Spaner D, Verulkar S, Mandal NP, Sinha PK, Peeraju P, Dongre PR, Mahto RN, et al. (2009) Characterization of the effect of a QTL for drought resistance in rice, qtl12.1, over a range of environments in the Philippines and eastern India. Euphytica 166: 207–217
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

