Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector
- PMID: 21602384
- PMCID: PMC3127684
- DOI: 10.1128/AEM.00417-11
Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector
Abstract
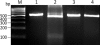
The oral spirochete Treponema denticola is associated with human periodontal disease. T. denticola ATCC 35405 and ATCC 33520 are two routinely used laboratory strains. Compared to T. denticola ATCC 33520, ATCC 35405 is more virulent but less accessible to genetic manipulations. For instance, the shuttle vectors of ATCC 33520 cannot be transformed into strain ATCC 35405. The lack of a shuttle vector has been a barrier to study the biology and virulence of T. denticola ATCC 35405. In this report, we hypothesize that T. denticola ATCC 35405 may have a unique DNA restriction-modification (R-M) system that prevents it from accepting the shuttle vectors of ATCC 33520 (e.g., the shuttle plasmid pBFC). To test this hypothesis, DNA restriction digestion, PCR, and Southern blot analyses were conducted to identify the differences between the R-M systems of these two strains. DNA restriction digestion analysis of these strains showed that only the cell extract from ATCC 35405 was able to digest pBFC. Consistently, PCR and Southern blot analyses revealed that the genome of T. denticola ATCC 35405 encodes three type II endonucleases that are absent in ATCC 33520. Among these three endonucleases, TDE0911 was predicted to cleave unmethylated double-stranded DNA and to be most likely responsible for the cleavage of unmethylated pBFC. In agreement with this prediction, the mutant of TDE0911 failed to cleave unmethylated pBFC plasmid, and it could accept the unmethylated shuttle vector. The study described here provides us with a new tool and strategy to genetically manipulate T. denticola, in particular ATCC 35405, and other strains that may carry similar endonucleases.
Figures


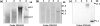
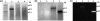


References
-
- Berndt C., Meier P., Wackernagel W. 2003. DNA restriction is a barrier to natural transformation in Pseudomonas stutzeri JM300. Microbiology 149: 895–901 - PubMed
-
- Capone R., et al. 2008. Human serum antibodies recognize Treponema denticola Msp and PrtP protease complex proteins. Oral Microbiol. Immunol. 23: 165–169 - PubMed
-
- Caudry S., et al. 1995. Distribution and characterization of plasmids in oral anaerobic spirochetes. Oral Microbiol. Immunol. 10: 8–12 - PubMed
-
- Chan E. C., et al. 1996. Characterization of a 4.2-kb plasmid isolated from periodontopathic spirochetes. Oral Microbiol. Immunol. 11: 365–368 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

