The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis
- PMID: 21602788
- PMCID: PMC3155295
- DOI: 10.1038/emboj.2011.158
The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis
Abstract
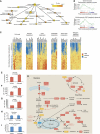
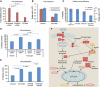
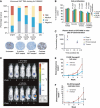
The androgen receptor (AR) is a key regulator of prostate growth and the principal drug target for the treatment of prostate cancer. Previous studies have mapped AR targets and identified some candidates which may contribute to cancer progression, but did not characterize AR biology in an integrated manner. In this study, we took an interdisciplinary approach, integrating detailed genomic studies with metabolomic profiling and identify an anabolic transcriptional network involving AR as the core regulator. Restricting flux through anabolic pathways is an attractive approach to deprive tumours of the building blocks needed to sustain tumour growth. Therefore, we searched for targets of the AR that may contribute to these anabolic processes and could be amenable to therapeutic intervention by virtue of differential expression in prostate tumours. This highlighted calcium/calmodulin-dependent protein kinase kinase 2, which we show is overexpressed in prostate cancer and regulates cancer cell growth via its unexpected role as a hormone-dependent modulator of anabolic metabolism. In conclusion, it is possible to progress from transcriptional studies to a promising therapeutic target by taking an unbiased interdisciplinary approach.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
From DNA binding to metabolic control: integration of -omics data reveals drug targets for prostate cancer.EMBO J. 2011 Jul 6;30(13):2516-7. doi: 10.1038/emboj.2011.205. EMBO J. 2011. PMID: 21731047 Free PMC article.
References
-
- Anderson KA, Ribar TJ, Lin F, Noeldner PK, Green MF, Muehlbauer MJ, Witters LA, Kemp BE, Means AR (2008) Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab 7: 377–388 - PubMed
-
- Attar RM, Jure-Kunkel M, Balog A, Cvijic ME, Dell-John J, Rizzo CA, Schweizer L, Spires TE, Platero JS, Obermeier M, Shan W, Salvati ME, Foster WR, Dinchuk J, Chen SJ, Vite G, Kramer R, Gottardis MM (2009) Discovery of BMS-641988, a novel and potent inhibitor of androgen receptor signaling for the treatment of prostate cancer. Cancer Res 69: 6522–6530 - PubMed
-
- Attard G, Reid AH, A'Hern R, Parker C, Oommen NB, Folkerd E, Messiou C, Molife LR, Maier G, Thompson E, Olmos D, Sinha R, Lee G, Dowsett M, Kaye SB, Dearnaley D, Kheoh T, Molina A, de Bono JS (2009) Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol 27: 3742–3748 - PMC - PubMed
-
- Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

