A cascade of protein kinase C isozymes promotes cytoskeletal polarization in T cells
- PMID: 21602810
- PMCID: PMC3119370
- DOI: 10.1038/ni.2033
A cascade of protein kinase C isozymes promotes cytoskeletal polarization in T cells
Abstract
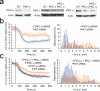
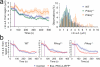
Polarization of the T cell microtubule-organizing center (MTOC) toward the antigen-presenting cell (APC) is driven by the accumulation of diacylglycerol (DAG) at the immunological synapse (IS). The mechanisms that couple DAG to the MTOC are not known. By single-cell photoactivation of the T cell antigen receptor (TCR), we found that three distinct isoforms of protein kinase C (PKC) were recruited by DAG to the IS in two steps. PKC-ɛ and PKC-η accumulated first in a broad region of membrane, whereas PKC-θ arrived later in a smaller zone. Functional experiments indicated that PKC-θ was required for MTOC reorientation and that PKC-ɛ and PKC-η operated redundantly to promote the recruitment of PKC-θ and subsequent polarization responses. Our results establish a previously uncharacterized role for PKC proteins in T cell polarity.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

