Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma
- PMID: 21602893
- PMCID: PMC3219832
- DOI: 10.1038/onc.2011.180
Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma
Abstract
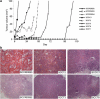
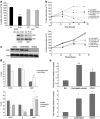
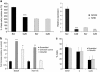
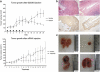
Identifying therapeutic targets for cancer treatment relies on consistent changes within particular types or sub-types of malignancy. The ability to define either consistent changes or sub-types of malignancy is often masked by tumor heterogeneity. To elucidate therapeutic targets in cutaneous squamous cell carcinoma (cSCC), the most frequent skin neoplasm with malignant potential, we have developed an integrated approach to gene expression profiling beginning with primary keratinocytes in culture. Candidate drivers of cSCC development were derived by first defining a set of in vitro cancer genes and then comparing their expression in a range of clinical data sets containing normal skin, cSCC and the benign hyper-proliferative condition psoriasis. A small interfering RNA (siRNA) screen of the resulting 21 upregulated genes has yielded targets capable of reducing xenograft tumor volume in vivo. Small-molecule inhibitors for one target, Polo-like kinase-1 (PLK1), are already in clinical trials for other malignancies, and our data show efficacy in cSCC. Another target, C20orf20, is identified as being overexpressed in cSCC, and siRNA-mediated knockdown induces apoptosis in vitro and reduces tumor growth in vivo. Thus, our approach has shown established and uncharacterized drivers of tumorigenesis with potent efficacy as therapeutic targets for the treatment of cSCC.
Figures

References
-
- Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. - PubMed
-
- Braakhuis BJ, Brakenhoff RH, Leemans CR. Gene expression profiling in head and neck squamous cell carcinoma. Curr Opin Otolaryngol Head Neck Surg. 2010;18:67–71. - PubMed
-
- Brattain MG, Brattain DE, Fine WD, Khaled FM, Marks ME, Kimball PM, et al. Initiation and characterization of cultures of human colonic carcinoma with different biological characteristics utilizing feeder layers of confluent fibroblasts. Oncodev Biol Med. 1981;2:355–366. - PubMed
-
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

