Ensemble and Single Molecule Studies on the Use of Metallic Nanostructures to Enhance the Intrinsic Emission of Enzyme Cofactors
- PMID: 21603075
- PMCID: PMC3097113
- DOI: 10.1021/jp112255j
Ensemble and Single Molecule Studies on the Use of Metallic Nanostructures to Enhance the Intrinsic Emission of Enzyme Cofactors
Abstract
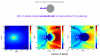
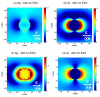
We present a strategy for enhancing the intrinsic emission of the enzyme cofactors flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN) and nicotinamide adenine dinucleotide (NADH). Ensemble studies show that silver island films (SIFs) are the optimal metal enhanced fluorescence (MEF) substrates for flavins and gave emission enhancements of over 10-fold for both FAD and FMN. A reduction in the lifetime of FAD and FMN on SIFs was also observed. Thermally evaporated aluminum films on quartz slides were found to be the optimal MEF substrate for NADH and gave a 5-fold increase in the emission intensity of NADH. We present finite-difference time-domain (FDTD) calculations that compute the enhancement in the radiated power emitting from an excited state dipole emitting in the wavelength range of NADH in close proximity to an aluminum nanoparticle, and a dipole emitting in the emission wavelength of flavins next to a silver nanoparticle. These calculations confirm that aluminum serves as the optimal MEF substrate for NADH and silver was the optimal MEF substrate for flavins. This is because the plasmon resonance properties of aluminum lie in the UV-blue regime and that of silver lie in the visible region. We also present the results of single molecule studies on FMN which show SIFs can both significantly enhance the intrinsic emission from single FMN molecules, significantly reduce their lifetimes and also significantly reduce FMN blinking. This is the first report of the observation of MEF from cofactors both at the ensemble and single molecule level. We hope this study will serve as a platform to encourage the future use of metallic nanostructures to study cofactors using their intrinsic fluorescence to directly monitor enzyme binding reactions without the need of extrinsic labeling of the molecules.
Figures







References
-
- Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Ed Springer; New York: 2006.
-
- Rigler R, Edman L, Földes-Papp Z, Wennmalm S. Fluorescence Correlation Spectroscopy in Single-Molecule Analysis: Enzymatic Catalysis at the Single Molecule Level. In: Rigler R, Orrit M, Basché T, editors. Single Molecule Spectroscopy - Nobel Conference Lectures. Springer; New York: 2001. p. 177.
-
- Visser AJWG, van den Berg PAW, Hink MA, Petushkov VN. Fluorescence Correlation Spectroscopy of Flavins and Flavoproteins. In: Rigler R, Elson ES, editors. Fluorescence Correlation Spectroscopy – Theory and Applications. Springer; New York: 2002. p. 9.
-
- van den Berg PAW, Visser AJWG. Tracking Molecular Dynamics of Flavoproteins with Time-Resolved Fluorescence Spectroscopy. In: Valeur B, Brochon B-V, editors. New Trends in Fluorescence Spectroscopy – Applications to Chemical and Life Sciences. Springer; New York: 2001. p. 457.
-
- van den Berg PAW, Widengren J, Hink MA, Rigler R, Visser AJWG. Spectrochim. Acta A. 2001;57:2135. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
