Platelets alter gene expression profile in human brain endothelial cells in an in vitro model of cerebral malaria
- PMID: 21603600
- PMCID: PMC3095604
- DOI: 10.1371/journal.pone.0019651
Platelets alter gene expression profile in human brain endothelial cells in an in vitro model of cerebral malaria
Abstract
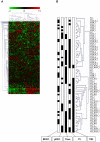
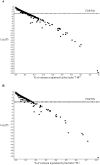
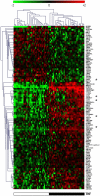
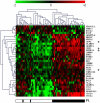
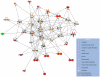
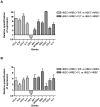
Platelet adhesion to the brain microvasculature has been associated with cerebral malaria (CM) in humans, suggesting that platelets play a role in the pathogenesis of this syndrome. In vitro co-cultures have shown that platelets can act as a bridge between Plasmodium falciparum-infected red blood cells (pRBC) and human brain microvascular endothelial cells (HBEC) and potentiate HBEC apoptosis. Using cDNA microarray technology, we analyzed transcriptional changes of HBEC in response to platelets in the presence or the absence of tumor necrosis factor (TNF) and pRBC, which have been reported to alter gene expression in endothelial cells. Using a rigorous statistical approach with multiple test corrections, we showed a significant effect of platelets on gene expression in HBEC. We also detected a strong effect of TNF, whereas there was no transcriptional change induced specifically by pRBC. Nevertheless, a global ANOVA and a two-way ANOVA suggested that pRBC acted in interaction with platelets and TNF to alter gene expression in HBEC. The expression of selected genes was validated by RT-qPCR. The analysis of gene functional annotation indicated that platelets induce the expression of genes involved in inflammation and apoptosis, such as genes involved in chemokine-, TREM1-, cytokine-, IL10-, TGFβ-, death-receptor-, and apoptosis-signaling. Overall, our results support the hypothesis that platelets play a pathogenic role in CM.
Conflict of interest statement
Figures
Similar articles
-
Platelets potentiate brain endothelial alterations induced by Plasmodium falciparum.Infect Immun. 2006 Jan;74(1):645-53. doi: 10.1128/IAI.74.1.645-653.2006. Infect Immun. 2006. PMID: 16369021 Free PMC article.
-
TGF-beta1 released from activated platelets can induce TNF-stimulated human brain endothelium apoptosis: a new mechanism for microvascular lesion during cerebral malaria.J Immunol. 2006 Jan 15;176(2):1180-4. doi: 10.4049/jimmunol.176.2.1180. J Immunol. 2006. PMID: 16394007
-
Plasmodium falciparum-infected erythrocytes induce tissue factor expression in endothelial cells and support the assembly of multimolecular coagulation complexes.J Thromb Haemost. 2007 Jan;5(1):155-65. doi: 10.1111/j.1538-7836.2006.02232.x. Epub 2006 Sep 26. J Thromb Haemost. 2007. PMID: 17002660 Free PMC article.
-
Platelet-endothelial cell interactions in cerebral malaria: the end of a cordial understanding.Thromb Haemost. 2009 Dec;102(6):1093-102. doi: 10.1160/TH09-05-0337. Thromb Haemost. 2009. PMID: 19967139 Review.
-
Cerebral malaria: role of microparticles and platelets in alterations of the blood-brain barrier.Int J Parasitol. 2006 May 1;36(5):541-6. doi: 10.1016/j.ijpara.2006.02.005. Epub 2006 Mar 10. Int J Parasitol. 2006. PMID: 16600245 Review.
Cited by
-
Cerebral Malaria and Neuronal Implications of Plasmodium Falciparum Infection: From Mechanisms to Advanced Models.Adv Sci (Weinh). 2022 Dec;9(36):e2202944. doi: 10.1002/advs.202202944. Epub 2022 Oct 27. Adv Sci (Weinh). 2022. PMID: 36300890 Free PMC article. Review.
-
Malaria: modification of the red blood cell and consequences in the human host.Br J Haematol. 2011 Sep;154(6):670-9. doi: 10.1111/j.1365-2141.2011.08755.x. Epub 2011 May 28. Br J Haematol. 2011. PMID: 21623767 Free PMC article. Review.
-
Platelet disturbances correlate with endothelial cell activation in uncomplicated Plasmodium vivax malaria.PLoS Negl Trop Dis. 2020 Jul 20;14(7):e0007656. doi: 10.1371/journal.pntd.0007656. eCollection 2020 Jul. PLoS Negl Trop Dis. 2020. PMID: 32687542 Free PMC article.
-
Experimental cerebral malaria pathogenesis--hemodynamics at the blood brain barrier.PLoS Pathog. 2014 Dec 4;10(12):e1004528. doi: 10.1371/journal.ppat.1004528. eCollection 2014 Dec. PLoS Pathog. 2014. PMID: 25474413 Free PMC article.
-
Endothelial cells potentiate interferon-γ production in a novel tripartite culture model of human cerebral malaria.PLoS One. 2013 Jul 12;8(7):e69521. doi: 10.1371/journal.pone.0069521. Print 2013. PLoS One. 2013. PMID: 23874969 Free PMC article.
References
-
- Marsh K, Snow RW. Malaria transmission and morbidity. Parassitologia. 1999;41:241–246. - PubMed
-
- van der Heyde HC, Nolan J, Combes V, Gramaglia I, Grau GE. A unified hypothesis for the genesis of cerebral malaria: sequestration, inflammation and hemostasis leading to microcirculatory dysfunction. Trends Parasitol. 2006;22:503–508. - PubMed
-
- Flori L, Delahaye NF, Iraqi FA, Hernandez-Valladares M, Fumoux F, et al. TNF as a malaria candidate gene: polymorphism-screening and family-based association analysis of mild malaria attack and parasitemia in Burkina Faso. Genes Immun. 2005;6:472–480. - PubMed
-
- Grau GE, Mackenzie CD, Carr RA, Redard M, Pizzolato G, et al. Platelet accumulation in brain microvessels in fatal pediatric cerebral malaria. J Infect Dis. 2003;187:461–466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

