Tau splicing and the intricacies of dementia
- PMID: 21604267
- PMCID: PMC3177961
- DOI: 10.1002/jcp.22842
Tau splicing and the intricacies of dementia
Abstract
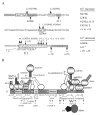
Tau is a microtubule-associated protein that fulfills several functions critical for neuronal formation and health. Tau discharges its functions by producing multiple isoforms via regulated alternative splicing. These isoforms modulate tau function in normal brain by altering the domains of the protein, thereby influencing its localization, conformation, and post-translational modifications and hence its availability and affinity for microtubules and other ligands. Disturbances in tau expression result in disruption of the neuronal cytoskeleton and formation of tau structures (neurofibrillary tangles) found in brains of dementia sufferers. More specifically, aberrations in tau splicing regulation directly cause several neurodegenerative diseases, which lead to dementia. In this review, I present our cumulative knowledge of tau splicing regulation in connection with neurodegeneration and also briefly go over the still-extensive list of questions that are connected to tau (dys)function.
Copyright © 2011 Wiley Periodicals, Inc.
Figures

References
-
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochem Biophys Acta. 2005;1739:91–103. - PubMed
-
- Andreadis A. Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog Mol Subcell Biol. 2006;44:89–107. - PubMed
-
- Brandt R, Hundelt M, Shahani N. Tau alteration and neuronal degeneration in tauopathies: mechanisms and models. Biochim Biophys Acta. 2005;1739:331–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

