Pharmer: efficient and exact pharmacophore search
- PMID: 21604800
- PMCID: PMC3124593
- DOI: 10.1021/ci200097m
Pharmer: efficient and exact pharmacophore search
Abstract
Pharmacophore search is a key component of many drug discovery efforts. Pharmer is a new computational approach to pharmacophore search that scales with the breadth and complexity of the query, not the size of the compound library being screened. Two novel methods for organizing pharmacophore data, the Pharmer KDB-tree and Bloom fingerprints, enable Pharmer to perform an exact pharmacophore search of almost two million structures in less than a minute. In general, Pharmer is more than an order of magnitude faster than existing technologies. The complete source code is available under an open-source license at http://pharmer.sourceforge.net .
Figures
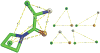





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

