A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages
- PMID: 21605306
- PMCID: PMC4185292
- DOI: 10.1111/j.1462-2920.2011.02502.x
A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages
Abstract
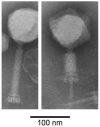
Bacteriophage S-CRM01 has been isolated from a freshwater strain of Synechococcus and shown to be present in the upper Klamath River valley in northern California and Oregon. The genome of this lytic T4-like phage has a 178,563 bp circular genetic map with 297 predicted protein-coding genes and 33 tRNA genes that represent all 20-amino-acid specificities. Analyses based on gene sequence and gene content indicate a close phylogenetic relationship to the 'photosynthetic' marine cyanomyophages infecting Synechococcus and Prochlorococcus. Such relatedness suggests that freshwater and marine phages can draw on a common gene pool. The genome can be considered as being comprised of three regions. Region 1 is populated predominantly with structural genes, recognized as such by homology to other T4-like phages and by identification in a proteomic analysis of purified virions. Region 2 contains most of the genes with roles in replication, recombination, nucleotide metabolism and regulation of gene expression, as well as 5 of the 6 signature genes of the photosynthetic cyanomyophages (hli03, hsp20, mazG, phoH and psbA; cobS is present in Region 3). Much of Regions 1 and 2 are syntenic with marine cyanomyophage genomes, except that a segment encompassing Region 2 is inverted. Region 3 contains a high proportion (85%) of genes that are unique to S-CRM01, as well as most of the tRNA genes. Regions 1 and 2 contain many predicted late promoters, with a combination of CTAAATA and ATAAATA core sequences. Two predicted genes that are unusual in phage genomes are homologues of cellular spoT and nusG.
© 2011 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures



Similar articles
-
Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges.Environ Microbiol. 2012 Feb;14(2):540-58. doi: 10.1111/j.1462-2920.2011.02667.x. Epub 2011 Dec 22. Environ Microbiol. 2012. PMID: 22188618
-
Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments.Environ Microbiol. 2010 Nov;12(11):3035-56. doi: 10.1111/j.1462-2920.2010.02280.x. Environ Microbiol. 2010. PMID: 20662890 Free PMC article.
-
Phylogenetic diversity of sequences of cyanophage photosynthetic gene psbA in marine and freshwaters.Appl Environ Microbiol. 2008 Sep;74(17):5317-24. doi: 10.1128/AEM.02480-07. Epub 2008 Jun 27. Appl Environ Microbiol. 2008. PMID: 18586962 Free PMC article.
-
Shedding new light on viral photosynthesis.Photosynth Res. 2015 Oct;126(1):71-97. doi: 10.1007/s11120-014-0057-x. Epub 2014 Nov 9. Photosynth Res. 2015. PMID: 25381655 Review.
-
T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology.Virol J. 2010 Oct 28;7:291. doi: 10.1186/1743-422X-7-291. Virol J. 2010. PMID: 21029435 Free PMC article. Review.
Cited by
-
A novel long-tailed myovirus represents a new T4-like cyanophage cluster.Front Microbiol. 2023 Nov 9;14:1293846. doi: 10.3389/fmicb.2023.1293846. eCollection 2023. Front Microbiol. 2023. PMID: 38029084 Free PMC article.
-
A New Freshwater Cyanosiphovirus Harboring Integrase.Front Microbiol. 2018 Sep 19;9:2204. doi: 10.3389/fmicb.2018.02204. eCollection 2018. Front Microbiol. 2018. PMID: 30283423 Free PMC article.
-
Prevalence of viral photosynthetic and capsid protein genes from cyanophages in two large and deep perialpine lakes.Appl Environ Microbiol. 2013 Dec;79(23):7169-78. doi: 10.1128/AEM.01914-13. Epub 2013 Sep 13. Appl Environ Microbiol. 2013. PMID: 24038692 Free PMC article.
-
Cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts.ISME J. 2012 Mar;6(3):619-28. doi: 10.1038/ismej.2011.146. Epub 2011 Oct 20. ISME J. 2012. PMID: 22011720 Free PMC article.
-
Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae.DNA Res. 2021 Aug 25;28(4):dsab013. doi: 10.1093/dnares/dsab013. DNA Res. 2021. PMID: 34390569 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

