Epigenetic factors and cardiac development
- PMID: 21606181
- PMCID: PMC3125076
- DOI: 10.1093/cvr/cvr138
Epigenetic factors and cardiac development
Abstract
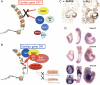
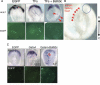
Congenital heart malformations remain the leading cause of death related to birth defects. Recent advances in developmental and regenerative cardiology have shed light on a mechanistic understanding of heart development that is controlled by a transcriptional network of genetic and epigenetic factors. This article reviews the roles of chromatin remodelling factors important for cardiac development with the current knowledge of cardiac morphogenesis, regeneration, and direct cardiac differentiation. In the last 5 years, critical roles of epigenetic factors have been revealed in the cardiac research field.
Figures


References
-
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi:10.1146/annurev.cellbio.23.090506.123331. - DOI - PubMed
-
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi:10.1038/nrg1710. - DOI - PubMed
-
- Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi:10.1016/j.cell.2006.11.031. - DOI - PubMed
-
- Moorman AFM, Webb S, Brown NA, Lamers W, Anderson RH. Development of the heart: (1) formation of the cardiac chambers and arterial trunks. Heart. 2003;89:806–814. doi:10.1136/heart.89.7.806. - DOI - PMC - PubMed
-
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi:10.1006/dbio.2000.9753. - DOI - PubMed

