Autoacetylation of the histone acetyltransferase Rtt109
- PMID: 21606491
- PMCID: PMC3137045
- DOI: 10.1074/jbc.M111.251579
Autoacetylation of the histone acetyltransferase Rtt109
Abstract
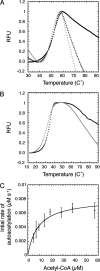
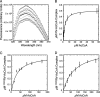
Rtt109 is a yeast histone acetyltransferase (HAT) that associates with histone chaperones Asf1 and Vps75 to acetylate H3K56, H3K9, and H3K27 and is important in DNA replication and maintaining genomic integrity. Recently, mass spectrometry and structural studies of Rtt109 have shown that active site residue Lys-290 is acetylated. However, the functional role of this modification and how the acetyl group is added to Lys-290 was unclear. Here, we examined the mechanism of Lys-290 acetylation and found that Rtt109 catalyzes intramolecular autoacetylation of Lys-290 ∼200-times slower than H3 acetylation. Deacetylated Rtt109 was prepared by reacting with a sirtuin protein deacetylase, producing an enzyme with negligible HAT activity. Autoacetylation of Rtt109 restored full HAT activity, indicating that autoacetylation is necessary for HAT activity and is a fully reversible process. To dissect the mechanism of activation, biochemical, and kinetic analyses were performed with Lys-290 variants of the Rtt109-Vps75 complex. We found that autoacetylation of Lys-290 increases the binding affinity for acetyl-CoA and enhances the rate of acetyl-transfer onto histone substrates. This study represents the first detailed investigation of a HAT enzyme regulated by single-site intramolecular autoacetylation.
Figures

References
-
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 - PubMed
-
- Spencer V. A., Davie J. R. (1999) Gene 240, 1–12 - PubMed
-
- Davie J. R. (1998) Curr. Opin. Genet. Dev. 8, 173–178 - PubMed
-
- Latham J. A., Dent S. Y. (2007) Nat. Struct. Mol. Biol. 14, 1017–1024 - PubMed
-
- Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

