Direct and indirect effects in the regulation of overlapping promoters
- PMID: 21609952
- PMCID: PMC3167631
- DOI: 10.1093/nar/gkr390
Direct and indirect effects in the regulation of overlapping promoters
Abstract
Optimal response to environmental stimuli often requires activation of certain genes and repression of others. Dual function regulatory proteins play a key role in the differential regulation of gene expression. While repression can be achieved by any DNA binding protein through steric occlusion of RNA polymerase in the promoter region, activation often requires a surface on the regulatory protein to contact RNAP and thus facilitate transcription initiation. RNAP itself is also a DNA binding protein, therefore it can function as a transcriptional repressor. Searching the Escherichia coli promoter database we found that ∼14% of the identified 'forward' promoters overlap with a promoter oriented in the opposite direction. In this article we combine a mathematical model with experimental analysis of synthetic regulatory regions to investigate interference of overlapping promoters. We find that promoter interference depends on the characteristics of overlapping promoters. The model predicts that promoter strength and interference can be regulated separately, which provides unique opportunities for regulation. Our experimental data suggest that in principle any DNA binding protein can be used for both activation and repression of promoter transcription, depending on the context. These findings can be exploited in the construction of synthetic networks.
Figures




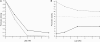
References
-
- Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002.
-
- Perez-Martin J, Espinosa M. Protein-induced bending as a transcriptional switch. Science. 1993;260:805–807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

