Depurination of Brome mosaic virus RNA3 inhibits its packaging into virus particles
- PMID: 21609957
- PMCID: PMC3167629
- DOI: 10.1093/nar/gkr383
Depurination of Brome mosaic virus RNA3 inhibits its packaging into virus particles
Abstract
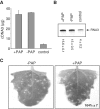
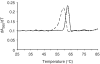
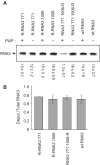
Packaging of the segmented RNA genome of Brome mosaic virus (BMV) into discrete particles is an essential step in the virus life cycle; however, questions remain regarding the mechanism of RNA packaging and the degree to which the viral coat protein controls the process. In this study, we used a plant-derived glycosidase, Pokeweed antiviral protein, to remove 14 specific bases from BMV RNA3 to examine the effect of depurination on virus assembly. Depurination of A771 within ORF3 and A1006 in the intergenic region inhibited coat protein binding and prevented RNA3 incorporation into particles. The disruption of interaction was not based on sequence identity, as mutation of these two purines to pyrimidines did not decrease coat protein-binding affinity. Rather, we suggest that base removal results in decreased thermodynamic stability of local RNA structures required for packaging, and that this instability is detected by coat protein. These results describe a new level of discrimination by coat protein, whereby it recognizes damage to specific viral RNA elements in the form of base removal and selects against incorporating the RNA into particles.
Figures







Similar articles
-
Depurination within the intergenic region of Brome mosaic virus RNA3 inhibits viral replication in vitro and in vivo.Nucleic Acids Res. 2008 Dec;36(22):7230-9. doi: 10.1093/nar/gkn896. Epub 2008 Nov 12. Nucleic Acids Res. 2008. PMID: 19004869 Free PMC article.
-
cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts.J Virol. 2003 Sep;77(18):9979-86. doi: 10.1128/jvi.77.18.9979-9986.2003. J Virol. 2003. PMID: 12941908 Free PMC article.
-
Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells.J Biol Chem. 2005 May 20;280(20):20069-75. doi: 10.1074/jbc.M413452200. Epub 2005 Mar 10. J Biol Chem. 2005. PMID: 15764597
-
The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein.Mol Plant Pathol. 2011 May;12(4):403-12. doi: 10.1111/j.1364-3703.2010.00678.x. Epub 2010 Nov 25. Mol Plant Pathol. 2011. PMID: 21453435 Free PMC article. Review.
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
Cited by
-
Toxic proteins in plants.Phytochemistry. 2015 Sep;117:51-64. doi: 10.1016/j.phytochem.2015.05.020. Epub 2015 Jun 7. Phytochemistry. 2015. PMID: 26057229 Free PMC article. Review.
-
Antiviral Activity of Ribosome-Inactivating Proteins.Toxins (Basel). 2021 Jan 22;13(2):80. doi: 10.3390/toxins13020080. Toxins (Basel). 2021. PMID: 33499086 Free PMC article. Review.
-
Ribosome depurination by ricin leads to inhibition of endoplasmic reticulum stress-induced HAC1 mRNA splicing on the ribosome.J Biol Chem. 2019 Nov 22;294(47):17848-17862. doi: 10.1074/jbc.RA119.009128. Epub 2019 Oct 17. J Biol Chem. 2019. PMID: 31624149 Free PMC article.
-
RIPpore: A Novel Host-Derived Method for the Identification of Ricin Intoxication through Oxford Nanopore Direct RNA Sequencing.Toxins (Basel). 2022 Jul 9;14(7):470. doi: 10.3390/toxins14070470. Toxins (Basel). 2022. PMID: 35878208 Free PMC article.
References
-
- Schmitz I, Rao AL. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–293. - PubMed
-
- Goldbach R, LeGall O, Wellink J. Alpha-like viruses in plants. Semin. Virol. 1991;2:19–25.
-
- Ahlquist P. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 1992;2:71–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

