Data mining using the Catalogue of Somatic Mutations in Cancer BioMart
- PMID: 21609966
- PMCID: PMC3263736
- DOI: 10.1093/database/bar018
Data mining using the Catalogue of Somatic Mutations in Cancer BioMart
Abstract
Catalogue of Somatic Mutations in Cancer (COSMIC) (http://www.sanger.ac.uk/cosmic) is a publicly available resource providing information on somatic mutations implicated in human cancer. Release v51 (January 2011) includes data from just over 19,000 genes, 161,787 coding mutations and 5573 gene fusions, described in more than 577,000 tumour samples. COSMICMart (COSMIC BioMart) provides a flexible way to mine these data and combine somatic mutations with other biological relevant data sets. This article describes the data available in COSMIC along with examples of how to successfully mine and integrate data sets using COSMICMart. DATABASE URL: http://www.sanger.ac.uk/genetics/CGP/cosmic/biomart/martview/.
Figures
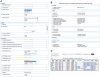
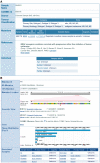
Similar articles
-
COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer.Nucleic Acids Res. 2011 Jan;39(Database issue):D945-50. doi: 10.1093/nar/gkq929. Epub 2010 Oct 15. Nucleic Acids Res. 2011. PMID: 20952405 Free PMC article.
-
COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer.Nucleic Acids Res. 2010 Jan;38(Database issue):D652-7. doi: 10.1093/nar/gkp995. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906727 Free PMC article.
-
COSMIC: High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer.Curr Protoc Hum Genet. 2016 Oct 11;91:10.11.1-10.11.37. doi: 10.1002/cphg.21. Curr Protoc Hum Genet. 2016. PMID: 27727438
-
The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers.Nat Rev Cancer. 2018 Nov;18(11):696-705. doi: 10.1038/s41568-018-0060-1. Nat Rev Cancer. 2018. PMID: 30293088 Free PMC article. Review.
-
Cytogenetic Resources and Information.Methods Mol Biol. 2017;1541:311-331. doi: 10.1007/978-1-4939-6703-2_25. Methods Mol Biol. 2017. PMID: 27910033 Review.
Cited by
-
Enabling a genetically informed approach to cancer medicine: a retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel.Oncologist. 2014 Jun;19(6):616-22. doi: 10.1634/theoncologist.2014-0011. Epub 2014 May 5. Oncologist. 2014. PMID: 24797823 Free PMC article.
-
Approaches to uncovering cancer diagnostic and prognostic molecular signatures.Mol Cell Oncol. 2014 Oct 29;1(2):e957981. doi: 10.4161/23723548.2014.957981. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308330 Free PMC article.
-
The BioMart community portal: an innovative alternative to large, centralized data repositories.Nucleic Acids Res. 2015 Jul 1;43(W1):W589-98. doi: 10.1093/nar/gkv350. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897122 Free PMC article.
-
Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours.PLoS One. 2011;6(12):e28250. doi: 10.1371/journal.pone.0028250. Epub 2011 Dec 6. PLoS One. 2011. PMID: 22163003 Free PMC article.
-
The Reactome BioMart.Database (Oxford). 2011 Oct 19;2011:bar031. doi: 10.1093/database/bar031. Print 2011. Database (Oxford). 2011. PMID: 22012987 Free PMC article.
References
-
- Den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. - PubMed
-
- Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

