Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation
- PMID: 21610030
- PMCID: PMC3100709
- DOI: 10.1242/dev.063479
Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation
Abstract
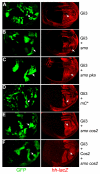
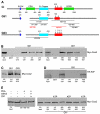
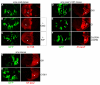
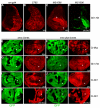
Hedgehog (Hh) signaling activates full-length Ci/Gli family transcription factors and prevents Ci/Gli proteolytic processing to repressor forms. In the absence of Hh, Ci/Gli processing is initiated by direct Pka phosphorylation. Despite those fundamental similarities between Drosophila and mammalian Hh pathways, the differential reliance on cilia and some key signal transduction components had suggested a major divergence in the mechanisms that regulate Ci/Gli protein activities, including the role of the kinesin-family protein Costal 2 (Cos2), which directs Ci processing in Drosophila. Here, we show that Cos2 binds to three regions of Gli1, just as for Ci, and that Cos2 functions to silence mammalian Gli1 in Drosophila in a Hh-regulated manner. Cos2 and the mammalian kinesin Kif7 can also direct Gli3 and Ci processing in fly, underscoring a fundamental conserved role for Cos2 family proteins in Hh signaling. We also show that direct PKA phosphorylation regulates the activity, rather than the proteolysis of Gli in Drosophilia, and we provide evidence for an analogous action of PKA on Ci.
Figures



Similar articles
-
Contributions of Costal 2-Fused interactions to Hedgehog signaling in Drosophila.Development. 2015 Mar 1;142(5):931-42. doi: 10.1242/dev.112904. Epub 2015 Jan 29. Development. 2015. PMID: 25633354 Free PMC article.
-
Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator.Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):E5651-60. doi: 10.1073/pnas.1416652111. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512501 Free PMC article.
-
Drosophila hedgehog can act as a morphogen in the absence of regulated Ci processing.Elife. 2020 Oct 21;9:e61083. doi: 10.7554/eLife.61083. Elife. 2020. PMID: 33084577 Free PMC article.
-
Regulation of Hh/Gli signaling by dual ubiquitin pathways.Cell Cycle. 2006 Nov 1;5(21):2457-63. doi: 10.4161/cc.5.21.3406. Epub 2006 Sep 14. Cell Cycle. 2006. PMID: 17102630 Review.
-
Gli protein nuclear localization signal.Vitam Horm. 2012;88:73-89. doi: 10.1016/B978-0-12-394622-5.00004-3. Vitam Horm. 2012. PMID: 22391300 Review.
Cited by
-
A mutation in the mouse ttc26 gene leads to impaired hedgehog signaling.PLoS Genet. 2014 Oct 23;10(10):e1004689. doi: 10.1371/journal.pgen.1004689. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340710 Free PMC article.
-
Hedgehog signaling regulates bladder cancer growth and tumorigenicity.Cancer Res. 2012 Sep 1;72(17):4449-58. doi: 10.1158/0008-5472.CAN-11-4123. Epub 2012 Jul 19. Cancer Res. 2012. PMID: 22815529 Free PMC article.
-
The cAMP effector PKA mediates Moody GPCR signaling in Drosophila blood-brain barrier formation and maturation.Elife. 2021 Aug 12;10:e68275. doi: 10.7554/eLife.68275. Elife. 2021. PMID: 34382936 Free PMC article.
-
Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo.PLoS Genet. 2013;9(12):e1003955. doi: 10.1371/journal.pgen.1003955. Epub 2013 Dec 5. PLoS Genet. 2013. PMID: 24339784 Free PMC article.
-
Decoding the phosphorylation code in Hedgehog signal transduction.Cell Res. 2013 Feb;23(2):186-200. doi: 10.1038/cr.2013.10. Epub 2013 Jan 22. Cell Res. 2013. PMID: 23337587 Free PMC article. Review.
References
-
- Aza-Blanc P., Lin H. Y., Ruiz i Altaba A., Kornberg T. B. (2000). Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development 127, 4293-4301. - PubMed
-
- Bai C. B., Joyner A. L. (2001). Gli1 can rescue the in vivo function of Gli2. Development 128, 5161-5172. - PubMed
-
- Barnfield P. C., Zhang X., Thanabalasingham V., Yoshida M., Hui C. C. (2005). Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation 73, 397-405. - PubMed
-
- Cheung H. O., Zhang X., Ribeiro A., Mo R., Makino S., Puviindran V., Law K. K., Briscoe J., Hui C. C. (2009). The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2, ra29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

