Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding
- PMID: 21610079
- PMCID: PMC3137057
- DOI: 10.1074/jbc.M111.247536
Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding
Abstract
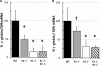
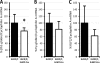
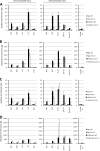
Krüppel-like factors (KLFs) control cell differentiation and embryonic development. KLF1 (erythroid Krüppel-like factor) plays essential roles in embryonic and adult erythropoiesis. KLF2 is a positive regulator of the mouse and human embryonic β-globin genes. KLF1 and KLF2 have highly homologous zinc finger DNA-binding domains. They have overlapping roles in embryonic erythropoiesis, as demonstrated using single and double KO mouse models. Ablation of the KLF1 or KLF2 gene causes embryonic lethality, but double KO embryos are more anemic and die sooner than either single KO. In this work, a dual human β-globin locus transgenic and KLF knockout mouse model was used. The results demonstrate that the human ε- (embryonic) and γ-globin (fetal) genes are positively regulated by KLF1 and KLF2 in embryos. Conditional KO mouse experiments indicate that the effect of KLF2 on embryonic globin gene regulation is at least partly erythroid cell-autonomous. KLF1 and KLF2 bind directly to the promoters of the human ε- and γ-globin genes, the mouse embryonic Ey- and βh1-globin genes, and also to the β-globin locus control region, as demonstrated by ChIP assays with mouse embryonic blood cells. H3K9Ac and H3K4me3 marks indicate open chromatin and active transcription, respectively. These marks are diminished at the Ey-, βh1-, ε- and γ-globin genes and locus control region in KLF1(-/-) embryos, correlating with reduced gene expression. Therefore, KLF1 and KLF2 positively regulate the embryonic and fetal β-globin genes through direct promoter binding. KLF1 is required for normal histone modifications in the β-globin locus in mouse embryos.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

